Evaluating a polymicrobial biofilm model for structural components by co-culturing Komagataeibacter hansenii produced bacterial cellulose with Pseudomonas aeruginosa PAO1
- PMID: 38322579
- PMCID: PMC10845243
- DOI: 10.1016/j.bioflm.2024.100176
Evaluating a polymicrobial biofilm model for structural components by co-culturing Komagataeibacter hansenii produced bacterial cellulose with Pseudomonas aeruginosa PAO1
Abstract
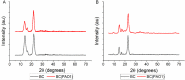
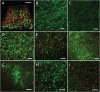
A polymicrobial biofilm model of Komagataeibacter hansenii and Pseudomonas aeruginosa was developed to understand whether a pre-existing matrix affects the ability of another species to build a biofilm. P. aeruginosa was inoculated onto the preformed K. hansenii biofilm consisting of a cellulose matrix. P. aeruginosa PAO1 colonized and infiltrated the K. hansenii bacterial cellulose biofilm (BC), as indicated by the presence of cells at 19 μm depth in the translucent hydrogel matrix. Bacterial cell density increased along the imaged depth of the biofilm (17-19 μm). On day 5, the average bacterial count across sections was 67 ± 4 % P. aeruginosa PAO1 and 33 ± 6 % K. hansenii. Biophysical characterization of the biofilm indicated that colonization by P. aeruginosa modified the biophysical properties of the BC matrix, which inlcuded increased density, heterogeneity, degradation temperature and thermal stability, and reduced crystallinity, swelling ability and moisture content. This further indicates colonization of the biofilm by P. aeruginosa. While eDNA fibres - a key viscoelastic component of P. aeruginosa biofilm - were present on the surface of the co-cultured biofilm on day 1, their abundance decreased over time, and by day 5, no eDNA was observed, either on the surface or within the matrix. P. aeruginosa-colonized biofilm devoid of eDNA retained its mechanical properties. The observations demonstrate that a pre-existing biofilm scaffold of K. hansenii inhibits P. aeruginosa PAO1 eDNA production and suggest that eDNA production is a response by P. aeruginosa to the viscoelastic properties of its environment.
Keywords: Biofilm; Co-culture; EPS; Extracellular DNA; Komagataeibacter hansenii; Microbial population; Pseudomonas aeruginosa PAO1; Structural integration.
© 2024 The Authors.
Conflict of interest statement
Authors have no competing interests in this study.
Figures






References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

