Structure-Activity Studies of Nitroreductase-Responsive Near-Infrared Heptamethine Cyanine Fluorescent Probes
- PMID: 38322783
- PMCID: PMC10846533
- DOI: 10.1002/ejoc.202200270
Structure-Activity Studies of Nitroreductase-Responsive Near-Infrared Heptamethine Cyanine Fluorescent Probes
Abstract
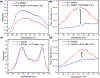
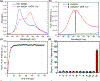
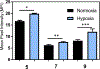
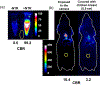
Two new classes of near-infrared molecular probes were prepared and shown to exhibit "turn on" fluorescence when cleaved by the nitroreductase enzyme, a well-known biomarker of cell hypoxia. The fluorescent probes are heptamethine cyanine dyes with a central 4'-carboxylic ester group on the heptamethine chain that is converted by a self-immolative fragmentation mechanism to a 4'-caboxylate group that greatly enhances the fluorescence brightness. Each compound was prepared by ring opening of a Zincke salt. The chemical structures have either terminal benzoindolinenes or propargyloxy auxochromes, which provide favorable red-shifted absorption/emission wavelengths and a hyperchromic effect that enhances the photon output when excited by 808 nm light. A fluorescent probe with terminal propargyloxy-indolenines exhibited less self-aggregation and was rapidly activated by nitroreductase with large "turn on" fluorescence; thus, it is the preferred choice for translation towards in vivo applications.
Keywords: NIR fluorescence; cell microscopy; cyanine dye; enzyme; molecular sensor.
Figures



References
-
- Abou Khouzam R; Brodaczewska K; Filipiak A; Zeinelabdin NA; Buart S; Szczylik C; Kieda C; Chouaib S Tumor Hypoxia Regulates Immune Escape/Invasion: Influence on Angiogenesis and Potential Impact of Hypoxic Biomarkers on Cancer Therapies. Front. Immunol 2021, 11, 1–16. 16.10.3389/fimmu.2020.613114. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
