All who wander are not lost: the search for homology during homologous recombination
- PMID: 38323621
- PMCID: PMC10903458
- DOI: 10.1042/BST20230705
All who wander are not lost: the search for homology during homologous recombination
Abstract
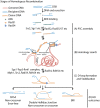
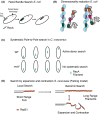
Homologous recombination (HR) is a template-based DNA double-strand break repair pathway that functions to maintain genomic integrity. A vital component of the HR reaction is the identification of template DNA to be used during repair. This occurs through a mechanism known as the homology search. The homology search occurs in two steps: a collision step in which two pieces of DNA are forced to collide and a selection step that results in homologous pairing between matching DNA sequences. Selection of a homologous template is facilitated by recombinases of the RecA/Rad51 family of proteins in cooperation with helicases, translocases, and topoisomerases that determine the overall fidelity of the match. This menagerie of molecular machines acts to regulate critical intermediates during the homology search. These intermediates include recombinase filaments that probe for short stretches of homology and early strand invasion intermediates in the form of displacement loops (D-loops) that stabilize paired DNA. Here, we will discuss recent advances in understanding how these specific intermediates are regulated on the molecular level during the HR reaction. We will also discuss how the stability of these intermediates influences the ultimate outcomes of the HR reaction. Finally, we will discuss recent physiological models developed to explain how the homology search protects the genome.
Keywords: DNA repair; Rad51; helicases; homologous recombination; homology search.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

