The probability of edge existence due to node degree: a baseline for network-based predictions
- PMID: 38323677
- PMCID: PMC10848215
- DOI: 10.1093/gigascience/giae001
The probability of edge existence due to node degree: a baseline for network-based predictions
Abstract
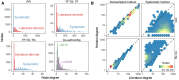
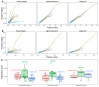
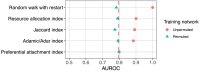
Important tasks in biomedical discovery such as predicting gene functions, gene-disease associations, and drug repurposing opportunities are often framed as network edge prediction. The number of edges connecting to a node, termed degree, can vary greatly across nodes in real biomedical networks, and the distribution of degrees varies between networks. If degree strongly influences edge prediction, then imbalance or bias in the distribution of degrees could lead to nonspecific or misleading predictions. We introduce a network permutation framework to quantify the effects of node degree on edge prediction. Our framework decomposes performance into the proportions attributable to degree and the network's specific connections using network permutation to generate features that depend only on degree. We discover that performance attributable to factors other than degree is often only a small portion of overall performance. Researchers seeking to predict new or missing edges in biological networks should use our permutation approach to obtain a baseline for performance that may be nonspecific because of degree. We released our methods as an open-source Python package (https://github.com/hetio/xswap/).
Keywords: Python; XSwap; bioinformatics; edge prediction; edge prior; heterogeneous; knowledge graphs; networks; node degree; permutation.
© The Author(s) 2024. Published by Oxford University Press GigaScience.
Conflict of interest statement
This work was supported, in part, by Pfizer Worldwide Research, Development, and Medical.
Figures






Update of
-
The probability of edge existence due to node degree: a baseline for network-based predictions.bioRxiv [Preprint]. 2023 Jan 6:2023.01.05.522939. doi: 10.1101/2023.01.05.522939. bioRxiv. 2023. Update in: Gigascience. 2024 Jan 2;13:giae001. doi: 10.1093/gigascience/giae001. PMID: 36711569 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

