Using blood transcriptome analysis for Alzheimer's disease diagnosis and patient stratification
- PMID: 38323937
- PMCID: PMC11032555
- DOI: 10.1002/alz.13691
Using blood transcriptome analysis for Alzheimer's disease diagnosis and patient stratification
Abstract
Introduction: Blood protein biomarkers demonstrate potential for Alzheimer's disease (AD) diagnosis. Limited studies examine the molecular changes in AD blood cells.
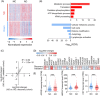
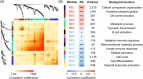
Methods: Bulk RNA-sequencing of blood cells was performed on AD patients of Chinese descent (n = 214 and 26 in the discovery and validation cohorts, respectively) with normal controls (n = 208 and 38 in the discovery and validation cohorts, respectively). Weighted gene co-expression network analysis (WGCNA) and deconvolution analysis identified AD-associated gene modules and blood cell types. Regression and unsupervised clustering analysis identified AD-associated genes, gene modules, cell types, and established AD classification models.
Results: WGCNA on differentially expressed genes revealed 15 gene modules, with 6 accurately classifying AD (areas under the receiver operating characteristics curve [auROCs] > 0.90). These modules stratified AD patients into subgroups with distinct disease states. Cell-type deconvolution analysis identified specific blood cell types potentially associated with AD pathogenesis.
Discussion: This study highlights the potential of blood transcriptome for AD diagnosis, patient stratification, and mechanistic studies.
Highlights: We comprehensively analyze the blood transcriptomes of a well-characterized Alzheimer's disease cohort to identify genes, gene modules, pathways, and specific blood cells associated with the disease. Blood transcriptome analysis accurately classifies and stratifies patients with Alzheimer's disease, with some gene modules achieving classification accuracy comparable to that of the plasma ATN biomarkers. Immune-associated pathways and immune cells, such as neutrophils, have potential roles in the pathogenesis and progression of Alzheimer's disease.
Keywords: Alzheimer's disease; blood; co‐expression; deconvolution; diagnosis; neutrophil; stratification; transcriptome.
© 2024 The Authors. Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Y.J., F.C.I., A.K.F., and N.Y.I. are inventors of the protein biomarker‐related technology licensed to Cognitact. Y.J. and F.C.I. are co‐founders of Cognitact. The remaining authors declare no competing interests. Author disclosures are available in the supporting information.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

