Exosomal circZNF800 Derived from Glioma Stem-like Cells Regulates Glioblastoma Tumorigenicity via the PIEZO1/Akt Axis
- PMID: 38324181
- PMCID: PMC11338982
- DOI: 10.1007/s12035-024-04002-0
Exosomal circZNF800 Derived from Glioma Stem-like Cells Regulates Glioblastoma Tumorigenicity via the PIEZO1/Akt Axis
Abstract
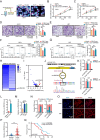
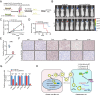
Exosomes play a crucial role in regulating crosstalk between tumor and tumor stem-like cells through their cargo molecules. Circular RNAs (circRNAs) have recently been demonstrated to be critical factors in tumorigenesis. This study focuses on the molecular mechanism by which circRNAs from glioma stem-like cell (GSLC) exosomes regulate glioblastoma (GBM) tumorigenicity. In this study, we validated that GSLC exosomes accelerated the malignant phenotype of GBM. Subsequently, we found that circZNF800 was highly expressed in GSLC exosomes and was negatively associated with GBM patients. CircZNF800 promoted GBM cell proliferation and migration and inhibited GBM cell apoptosis in vitro. Silencing circZNF800 could improve the GBM xenograft model survival rate. Mechanistic studies revealed that circZNF800 activated the PIEZO1/Akt signaling pathway by sponging miR-139-5p. CircZNF800 derived from GSLC exosomes promoted GBM cell tumorigenicity and predicted poor prognosis in GBM patients. CircZNF800 has the potential to serve as a promising target for further therapeutic exploration.
Keywords: Exosome; Glioma stem-like cell; PIEZO1; circZNF800; miR-139-5p.
© 2024. The Author(s).
Conflict of interest statement
The authors declare there are no potential competing interests.
Figures



Similar articles
-
Glioma-Associated Mesenchymal Stromal/Stem Cells Derived Exosomal miR-191 Promotes the Proneural-to-Mesenchymal Transition in Glioblastoma Cells via PTEN/PI3K/AKT Signaling.Int J Nanomedicine. 2025 Jul 7;20:8811-8831. doi: 10.2147/IJN.S515771. eCollection 2025. Int J Nanomedicine. 2025. PMID: 40657483 Free PMC article.
-
METTL8 links mt-tRNA m3C modification to the HIF1α/RTK/Akt axis to sustain GBM stemness and tumorigenicity.Cell Death Dis. 2024 May 14;15(5):338. doi: 10.1038/s41419-024-06718-2. Cell Death Dis. 2024. PMID: 38744809 Free PMC article.
-
Human bone marrow-derived mesenchymal stem cell-secreted exosomes overexpressing microRNA-34a ameliorate glioblastoma development via down-regulating MYCN.Cell Oncol (Dordr). 2019 Dec;42(6):783-799. doi: 10.1007/s13402-019-00461-z. Epub 2019 Jul 22. Cell Oncol (Dordr). 2019. Retraction in: Cell Oncol (Dordr). 2021 Oct;44(5):1207. doi: 10.1007/s13402-021-00627-8. PMID: 31332647 Retracted.
-
Integrative analysis of cell adhesion molecules in glioblastoma identified prostaglandin F2 receptor inhibitor (PTGFRN) as an essential gene.BMC Cancer. 2022 Jun 11;22(1):642. doi: 10.1186/s12885-022-09682-2. BMC Cancer. 2022. PMID: 35690717 Free PMC article. Review.
-
Molecular Mechanisms and Strategies for Inducing Neuronal Differentiation in Glioblastoma Cells.Cell Reprogram. 2025 Feb;27(1):24-32. doi: 10.1089/cell.2024.0087. Epub 2025 Jan 30. Cell Reprogram. 2025. PMID: 39880036 Review.
Cited by
-
The dual role of Piezo1 in tumor cells and immune cells: a new target for cancer therapy.Front Immunol. 2025 Jul 31;16:1635388. doi: 10.3389/fimmu.2025.1635388. eCollection 2025. Front Immunol. 2025. PMID: 40821847 Free PMC article. Review.
-
Deciphering the Role of Functional Ion Channels in Cancer Stem Cells (CSCs) and Their Therapeutic Implications.Int J Mol Sci. 2025 Aug 6;26(15):7595. doi: 10.3390/ijms26157595. Int J Mol Sci. 2025. PMID: 40806720 Free PMC article. Review.
-
Exploring the roles and clinical potential of exosome-derived non-coding RNAs in glioma.IBRO Neurosci Rep. 2025 Feb 5;18:323-337. doi: 10.1016/j.ibneur.2025.01.015. eCollection 2025 Jun. IBRO Neurosci Rep. 2025. PMID: 40034544 Free PMC article. Review.
-
Piezo1-related physiological and pathological processes in glioblastoma.Front Cell Dev Biol. 2025 Feb 21;13:1536320. doi: 10.3389/fcell.2025.1536320. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40061015 Free PMC article. Review.
-
Piezo1 in microglial cells: Implications for neuroinflammation and tumorigenesis.Channels (Austin). 2025 Dec;19(1):2492161. doi: 10.1080/19336950.2025.2492161. Epub 2025 Apr 13. Channels (Austin). 2025. PMID: 40223276 Free PMC article. Review.
References
-
- Meyer MA (2008) Malignant gliomas in adults. N Engl J Med 359(17):1850 author reply 1850 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

