Concurrent Clade I and Clade II Monkeypox Virus Circulation, Cameroon, 1979-2022
- PMID: 38325363
- PMCID: PMC10902553
- DOI: 10.3201/eid3003.230861
Concurrent Clade I and Clade II Monkeypox Virus Circulation, Cameroon, 1979-2022
Abstract
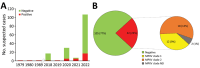
During 1979-2022, Cameroon recorded 32 laboratory-confirmed mpox cases among 137 suspected mpox cases identified by the national surveillance network. The highest positivity rate occurred in 2022, indicating potential mpox re-emergence in Cameroon. Both clade I (n = 12) and clade II (n = 18) monkeypox virus (MPXV) were reported, a unique feature of mpox in Cameroon. The overall case-fatality ratio of 2.2% was associated with clade II. We found mpox occurred only in the forested southern part of the country, and MPXV phylogeographic structure revealed a clear geographic separation among concurrent circulating clades. Clade I originated from eastern regions close to neighboring mpox-endemic countries in Central Africa; clade II was prevalent in western regions close to West Africa. Our findings suggest that MPXV re-emerged after a 30-year lapse and might arise from different viral reservoirs unique to ecosystems in eastern and western rainforests of Cameroon.
Keywords: Cameroon; Central Africa; MPXV; Mpox; West Africa; clade I; clade II; concurrent circulation; epidemiology; forest; geographic segregation; monkeypox virus; surveillance; surveillance system; viruses; zoonoses.
Figures



References
-
- Marennikova SS, Moyer RW. Classification of poxviruses and brief characterization of the genus Orthopoxvirus. In: Shchelkunov SN, Marennikova SS, Moyer RW, editors. Orthopoxviruses pathogenic for humans. Boston: Springer; 2005. p. 11–8.
-
- von Magnus P, Andersen EK, Petersen KB, Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Pathol Microbiol Scand. 1959;46:156–76. 10.1111/j.1699-0463.1959.tb00328.x - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

