The Mutographs biorepository: A unique genomic resource to study cancer around the world
- PMID: 38325367
- PMCID: PMC10943582
- DOI: 10.1016/j.xgen.2024.100500
The Mutographs biorepository: A unique genomic resource to study cancer around the world
Abstract
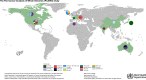
Large-scale biorepositories and databases are essential to generate equitable, effective, and sustainable advances in cancer prevention, early detection, cancer therapy, cancer care, and surveillance. The Mutographs project has created a large genomic dataset and biorepository of over 7,800 cancer cases from 30 countries across five continents with extensive demographic, lifestyle, environmental, and clinical information. Whole-genome sequencing is being finalized for over 4,000 cases, with the primary goal of understanding the causes of cancer at eight anatomic sites. Genomic, exposure, and clinical data will be publicly available through the International Cancer Genome Consortium Accelerating Research in Genomic Oncology platform. The Mutographs sample and metadata biorepository constitutes a legacy resource for new projects and collaborations aiming to increase our current research efforts in cancer genomic epidemiology globally.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests M.R.S. is founder of, consultant to, and stockholder in Quotient Therapeutics. L.B.A. is a compensated consultant and has equity interest in io9, LLC, and Genome Insight. His spouse is an employee of Biotheranostics, Inc. L.B.A. is also an inventor of a US patent 10,776,718 for source identification by non-negative matrix factorization. L.B.A. declares US provisional applications with serial numbers 63/289,601; 63/269,033; and 63/483,237. L.B.A. also declares US provisional applications with serial numbers 63/366,392; 63/367,846; 63/412,835; and 63/492,348.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

