An alternative splicing signature defines the basal-like phenotype and predicts worse clinical outcome in pancreatic cancer
- PMID: 38325381
- PMCID: PMC10897606
- DOI: 10.1016/j.xcrm.2024.101411
An alternative splicing signature defines the basal-like phenotype and predicts worse clinical outcome in pancreatic cancer
Abstract
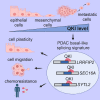
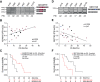
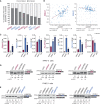
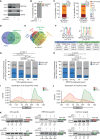
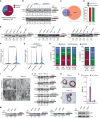
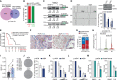
Pancreatic ductal adenocarcinoma (PDAC) is characterized by extremely poor prognosis. PDAC presents with molecularly distinct subtypes, with the basal-like one being associated with enhanced chemoresistance. Splicing dysregulation contributes to PDAC; however, its involvement in subtype specification remains elusive. Herein, we uncover a subtype-specific splicing signature associated with prognosis in PDAC and the splicing factor Quaking (QKI) as a determinant of the basal-like signature. Single-cell sequencing analyses highlight QKI as a marker of the basal-like phenotype. QKI represses splicing events associated with the classical subtype while promoting basal-like events associated with shorter survival. QKI favors a plastic, quasi-mesenchymal phenotype that supports migration and chemoresistance in PDAC organoids and cell lines, and its expression is elevated in high-grade primary tumors and metastatic lesions. These studies identify a splicing signature that defines PDAC subtypes and indicate that QKI promotes an undifferentiated, plastic phenotype, which renders PDAC cells chemoresistant and adaptable to environmental changes.
Keywords: RNA processing; alternative splicing; chemoresistance.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Neoptolemos J.P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018;15:333–348. - PubMed
-
- Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395:2008–2020. - PubMed
-
- Javed M.A., Beyer G., Le N., Vinci A., Wong H., Palmer D., Morgan R.D., Lamarca A., Hubner R.A., Valle J.W., et al. Impact of intensified chemotherapy in metastatic pancreatic ductal adenocarcinoma (PDAC) in clinical routine in Europe. Pancreatology. 2019;19:97–104. - PubMed
-
- Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16:207–220. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

