The molecular language of RNA 5' ends: guardians of RNA identity and immunity
- PMID: 38325897
- PMCID: PMC10946433
- DOI: 10.1261/rna.079942.124
The molecular language of RNA 5' ends: guardians of RNA identity and immunity
Abstract
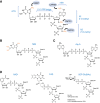
RNA caps are deposited at the 5' end of RNA polymerase II transcripts. This modification regulates several steps of gene expression, in addition to marking transcripts as self to enable the innate immune system to distinguish them from uncapped foreign RNAs, including those derived from viruses. Specialized immune sensors, such as RIG-I and IFITs, trigger antiviral responses upon recognition of uncapped cytoplasmic transcripts. Interestingly, uncapped transcripts can also be produced by mammalian hosts. For instance, 5'-triphosphate RNAs are generated by RNA polymerase III transcription, including tRNAs, Alu RNAs, or vault RNAs. These RNAs have emerged as key players of innate immunity, as they can be recognized by the antiviral sensors. Mechanisms that regulate the presence of 5'-triphosphates, such as 5'-end dephosphorylation or RNA editing, prevent immune recognition of endogenous RNAs and excessive inflammation. Here, we provide a comprehensive overview of the complexity of RNA cap structures and 5'-triphosphate RNAs, highlighting their roles in transcript identity, immune surveillance, and disease.
Keywords: 5′ cap; CMTR1; CMTR2; IFIT; RIG-I; antiviral; capping; m7G; type I interferon.
© 2024 Avila-Bonilla and Macias; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

Similar articles
-
Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.Proc Natl Acad Sci U S A. 2016 Jan 19;113(3):596-601. doi: 10.1073/pnas.1515152113. Epub 2016 Jan 5. Proc Natl Acad Sci U S A. 2016. PMID: 26733676 Free PMC article.
-
RIG-I recognizes metabolite-capped RNAs as signaling ligands.Nucleic Acids Res. 2023 Aug 25;51(15):8102-8114. doi: 10.1093/nar/gkad518. Nucleic Acids Res. 2023. PMID: 37326006 Free PMC article.
-
Characterization of rotavirus RNAs that activate innate immune signaling through the RIG-I-like receptors.PLoS One. 2013 Jul 23;8(7):e69825. doi: 10.1371/journal.pone.0069825. Print 2013. PLoS One. 2013. PMID: 23894547 Free PMC article.
-
The RNA polymerase III-RIG-I axis in antiviral immunity and inflammation.Trends Immunol. 2023 Jun;44(6):435-449. doi: 10.1016/j.it.2023.04.002. Epub 2023 May 4. Trends Immunol. 2023. PMID: 37149405 Free PMC article. Review.
-
In vitro production of synthetic viral RNAs and their delivery into mammalian cells and the application of viral RNAs in the study of innate interferon responses.Methods. 2020 Nov 1;183:21-29. doi: 10.1016/j.ymeth.2019.10.013. Epub 2019 Nov 1. Methods. 2020. PMID: 31682923 Review.
Cited by
-
5' DREDGE: Direct Repeat-Enabled Downregulation of Gene Expression via the 5' UTR of Target Genes.Cells. 2025 Jun 8;14(12):866. doi: 10.3390/cells14120866. Cells. 2025. PMID: 40558493 Free PMC article.
-
A tRNA gene potential to activate interferon signaling involves selective termination and is suppressible by La protein/SSB.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf513. doi: 10.1093/nar/gkaf513. Nucleic Acids Res. 2025. PMID: 40637226 Free PMC article.
-
The Role of Chemical Modifications in the Genome of Negative-Sense RNA Viruses on the Innate Immune Response.Viruses. 2025 May 30;17(6):795. doi: 10.3390/v17060795. Viruses. 2025. PMID: 40573386 Free PMC article. Review.
-
Structure-Based Identification of SARS-CoV-2 nsp10-16 Methyltransferase Inhibitors Using Molecular Dynamics Insights.Curr Issues Mol Biol. 2025 Mar 17;47(3):198. doi: 10.3390/cimb47030198. Curr Issues Mol Biol. 2025. PMID: 40136452 Free PMC article.
-
Multi-omics analysis reveals CMTR1 upregulation in cancer and roles in ribosomal protein gene expression and tumor growth.Cell Commun Signal. 2025 Apr 24;23(1):197. doi: 10.1186/s12964-025-02147-6. Cell Commun Signal. 2025. PMID: 40275371 Free PMC article.
References
-
- Abbas YM, Laudenbach BT, Martínez-Montero S, Cencic R, Habjan M, Pichlmair A, Damha MJ, Pelletier J, Nagar B. 2017. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc Natl Acad Sci 114: E2106–E2115. 10.1073/pnas.1612444114 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
