Structural basis for the oligomerization-facilitated NLRP3 activation
- PMID: 38326375
- PMCID: PMC10850481
- DOI: 10.1038/s41467-024-45396-8
Structural basis for the oligomerization-facilitated NLRP3 activation
Abstract
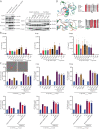
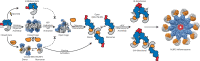
The NACHT-, leucine-rich-repeat-, and pyrin domain-containing protein 3 (NLRP3) is a critical intracellular inflammasome sensor and an important clinical target against inflammation-driven human diseases. Recent studies have elucidated its transition from a closed cage to an activated disk-like inflammasome, but the intermediate activation mechanism remains elusive. Here we report the cryo-electron microscopy structure of NLRP3, which forms an open octamer and undergoes a ~ 90° hinge rotation at the NACHT domain. Mutations on open octamer's interfaces reduce IL-1β signaling, highlighting its essential role in NLRP3 activation/inflammasome assembly. The centrosomal NIMA-related kinase 7 (NEK7) disrupts large NLRP3 oligomers and forms NEK7/NLRP3 monomers/dimers which is a critical step preceding the assembly of the disk-like inflammasome. These data demonstrate an oligomeric cooperative activation of NLRP3 and provide insight into its inflammasome assembly mechanism.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: X.Y., R.Ma., R.Mi., D.C., B.V.S., K.G., J.S., B.P., Y.Y., G.J.T., L.P., A.B., D.O., N.V.O., and S.S. are employees of Johnson & Johnson Innovative Medicine. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

