Genome-wide association study identifies human genetic variants associated with fatal outcome from Lassa fever
- PMID: 38326571
- PMCID: PMC10914620
- DOI: 10.1038/s41564-023-01589-3
Genome-wide association study identifies human genetic variants associated with fatal outcome from Lassa fever
Abstract
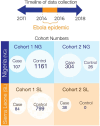
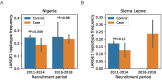
Infection with Lassa virus (LASV) can cause Lassa fever, a haemorrhagic illness with an estimated fatality rate of 29.7%, but causes no or mild symptoms in many individuals. Here, to investigate whether human genetic variation underlies the heterogeneity of LASV infection, we carried out genome-wide association studies (GWAS) as well as seroprevalence surveys, human leukocyte antigen typing and high-throughput variant functional characterization assays. We analysed Lassa fever susceptibility and fatal outcomes in 533 cases of Lassa fever and 1,986 population controls recruited over a 7 year period in Nigeria and Sierra Leone. We detected genome-wide significant variant associations with Lassa fever fatal outcomes near GRM7 and LIF in the Nigerian cohort. We also show that a haplotype bearing signatures of positive selection and overlapping LARGE1, a required LASV entry factor, is associated with decreased risk of Lassa fever in the Nigerian cohort but not in the Sierra Leone cohort. Overall, we identified variants and genes that may impact the risk of severe Lassa fever, demonstrating how GWAS can provide insight into viral pathogenesis.
© 2024. The Author(s).
Conflict of interest statement
P.C.S., R. Tewhey and S.K.R. are inventors on patents related to massively parallel reporter assays. P.C.S. is a co-founder of, shareholder in and consultant to Sherlock Biosciences, Inc. and Delve Bio, as well as a Board member of and shareholder in Danaher Corporation. The other authors declare no competing interests.
Figures



References
-
- McCormick JB, Fisher-Hoch SP. Lassa fever. Curr. Top. Microbiol. Immunol. 2002;262:75–109. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

