Identification of direct connections between the dura and the brain
- PMID: 38326613
- PMCID: PMC11254388
- DOI: 10.1038/s41586-023-06993-7
Identification of direct connections between the dura and the brain
Abstract
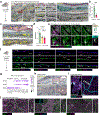
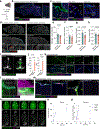
The arachnoid barrier delineates the border between the central nervous system and dura mater. Although the arachnoid barrier creates a partition, communication between the central nervous system and the dura mater is crucial for waste clearance and immune surveillance1,2. How the arachnoid barrier balances separation and communication is poorly understood. Here, using transcriptomic data, we developed transgenic mice to examine specific anatomical structures that function as routes across the arachnoid barrier. Bridging veins create discontinuities where they cross the arachnoid barrier, forming structures that we termed arachnoid cuff exit (ACE) points. The openings that ACE points create allow the exchange of fluids and molecules between the subarachnoid space and the dura, enabling the drainage of cerebrospinal fluid and limited entry of molecules from the dura to the subarachnoid space. In healthy human volunteers, magnetic resonance imaging tracers transit along bridging veins in a similar manner to access the subarachnoid space. Notably, in neuroinflammatory conditions such as experimental autoimmune encephalomyelitis, ACE points also enable cellular trafficking, representing a route for immune cells to directly enter the subarachnoid space from the dura mater. Collectively, our results indicate that ACE points are a critical part of the anatomy of neuroimmune communication in both mice and humans that link the central nervous system with the dura and its immunological diversity and waste clearance systems.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures













References
Reference list
Additional references
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

