Naturally occurring T cell mutations enhance engineered T cell therapies
- PMID: 38326614
- PMCID: PMC11573425
- DOI: 10.1038/s41586-024-07018-7
Naturally occurring T cell mutations enhance engineered T cell therapies
Abstract
Adoptive T cell therapies have produced exceptional responses in a subset of patients with cancer. However, therapeutic efficacy can be hindered by poor T cell persistence and function1. In human T cell cancers, evolution of the disease positively selects for mutations that improve fitness of T cells in challenging situations analogous to those faced by therapeutic T cells. Therefore, we reasoned that these mutations could be co-opted to improve T cell therapies. Here we systematically screened the effects of 71 mutations from T cell neoplasms on T cell signalling, cytokine production and in vivo persistence in tumours. We identify a gene fusion, CARD11-PIK3R3, found in a CD4+ cutaneous T cell lymphoma2, that augments CARD11-BCL10-MALT1 complex signalling and anti-tumour efficacy of therapeutic T cells in several immunotherapy-refractory models in an antigen-dependent manner. Underscoring its potential to be deployed safely, CARD11-PIK3R3-expressing cells were followed up to 418 days after T cell transfer in vivo without evidence of malignant transformation. Collectively, our results indicate that exploiting naturally occurring mutations represents a promising approach to explore the extremes of T cell biology and discover how solutions derived from evolution of malignant T cells can improve a broad range of T cell therapies.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures










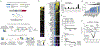
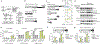
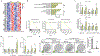
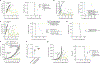

Comment in
-
Increasing the potency of T cell therapies.Nat Rev Drug Discov. 2024 Apr;23(4):253. doi: 10.1038/d41573-024-00043-1. Nat Rev Drug Discov. 2024. PMID: 38448667 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

