Spatial transcriptomics reveal neuron-astrocyte synergy in long-term memory
- PMID: 38326616
- PMCID: PMC10937396
- DOI: 10.1038/s41586-023-07011-6
Spatial transcriptomics reveal neuron-astrocyte synergy in long-term memory
Abstract
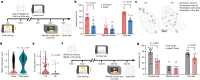
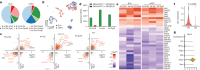
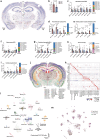
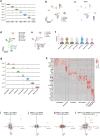
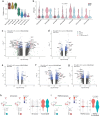
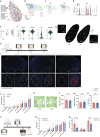
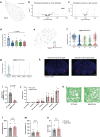
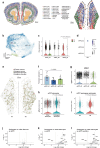
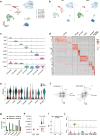
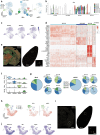
Memory encodes past experiences, thereby enabling future plans. The basolateral amygdala is a centre of salience networks that underlie emotional experiences and thus has a key role in long-term fear memory formation1. Here we used spatial and single-cell transcriptomics to illuminate the cellular and molecular architecture of the role of the basolateral amygdala in long-term memory. We identified transcriptional signatures in subpopulations of neurons and astrocytes that were memory-specific and persisted for weeks. These transcriptional signatures implicate neuropeptide and BDNF signalling, MAPK and CREB activation, ubiquitination pathways, and synaptic connectivity as key components of long-term memory. Notably, upon long-term memory formation, a neuronal subpopulation defined by increased Penk and decreased Tac expression constituted the most prominent component of the memory engram of the basolateral amygdala. These transcriptional changes were observed both with single-cell RNA sequencing and with single-molecule spatial transcriptomics in intact slices, thereby providing a rich spatial map of a memory engram. The spatial data enabled us to determine that this neuronal subpopulation interacts with adjacent astrocytes, and functional experiments show that neurons require interactions with astrocytes to encode long-term memory.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Comment in
-
False positives in study of memory-related gene expression.Nature. 2025 Jun;642(8066):E1-E3. doi: 10.1038/s41586-025-08988-y. Epub 2025 Jun 4. Nature. 2025. PMID: 40467748 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

