RNA-binding protein RPS7 promotes hepatocellular carcinoma progression via LOXL2-dependent activation of ITGB1/FAK/SRC signaling
- PMID: 38326908
- PMCID: PMC10851485
- DOI: 10.1186/s13046-023-02929-1
RNA-binding protein RPS7 promotes hepatocellular carcinoma progression via LOXL2-dependent activation of ITGB1/FAK/SRC signaling
Erratum in
-
Correction: RNA-binding protein RPS7 promotes hepatocellular carcinoma progression via LOXL2-dependent activation of ITGB1/FAK/SRC signaling.J Exp Clin Cancer Res. 2024 Mar 14;43(1):80. doi: 10.1186/s13046-024-02999-9. J Exp Clin Cancer Res. 2024. PMID: 38481346 Free PMC article. No abstract available.
Abstract
Background: Metastasis is one of the leading cause contributes to treatment failure and poor prognosis of hepatocellular carcinoma (HCC) patients. The underlying mechanism of HCC metastasis remains to be determined. Although several RNA binding proteins (RBPs) have been found to participate in tumorigenesis and progression of liver cancer, the role of RBPs in HCC patients with extrahepatic metastases is poorly understood.
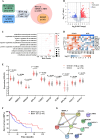
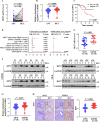
Methods: By performing RNA-seq of primary HCC tissues (including HCC with extrahepatic metastasis and those did not develop metastasis), we identified a set of HCC metastasis-associated RBPs candidates. Among which, ribosomal protein S7 (RPS7) was found to be remarkably increased in HCC tissues and be strongly related to HCC poor survival. Overexpression or CRISPR-Cas9-mediated knockout were applied to investigate the role of RPS7 on the metastasis-associated phenotypes of HCC cells. RNA sequencing, RIP, RNA-pull down, dual luciferase reporter assay, nascent RNA capture assay, and RNA decay and so on, were applied to reveal the underlying mechanism of RPS7 induced HCC metastasis.
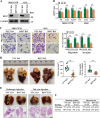
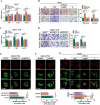
Results: Gain- and loss- of function analyses revealed that RPS7 promoted HCC cells adhesion, migration and invasion capabilities, as well as lung metastasis. Mechanistically, we uncovered that lysyl oxidase-like 2 (LOXL2) was a critical downstream target of RPS7. RPS7 could stabilize LOXL2 mRNA by binding to AUUUA motifs in the 3155-3375 region of the 3'UTR of LOXL2 mRNA, thus increased LOXL2 expression via elevating LOXL2 mRNA abundance. Further research revealed that LOXL2 could accelerate focal adhesion formation through maintaining the protein stability of ITGB1 and activating ITGB1-mediated FAK/SRC signaling pathway, and thereby contribute to the pro-metastasis effect of RPS7.
Conclusions: Taken together, our data reveal a novel function of RPS7 in HCC metastasis, also reveal the critical roles of the RPS7/LOXL2/ITGB1 axis in HCC metastasis and shed new light on the exploration of molecular drugs against HCC.
Keywords: Focal adhesion; Hepatocellular carcinoma; Lysyl oxidase-like 2; Metastasis; RNA-binding protein; Ribosomal protein S7.
© 2024. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

