This is a preprint.
Mouse Adaptation of Human Inflammatory Bowel Diseases Microbiota Enhances Colonization Efficiency and Alters Microbiome Aggressiveness Depending on Recipient Colonic Inflammatory Environment
- PMID: 38328082
- PMCID: PMC10849574
- DOI: 10.1101/2024.01.23.576862
Mouse Adaptation of Human Inflammatory Bowel Diseases Microbiota Enhances Colonization Efficiency and Alters Microbiome Aggressiveness Depending on Recipient Colonic Inflammatory Environment
Update in
-
Mouse adaptation of human inflammatory bowel diseases microbiota enhances colonization efficiency and alters microbiome aggressiveness depending on the recipient colonic inflammatory environment.Microbiome. 2024 Aug 7;12(1):147. doi: 10.1186/s40168-024-01857-2. Microbiome. 2024. PMID: 39113097 Free PMC article.
Abstract
Understanding the cause vs consequence relationship of gut inflammation and microbial dysbiosis in inflammatory bowel diseases (IBD) requires a reproducible mouse model of human-microbiota-driven experimental colitis. Our study demonstrated that human fecal microbiota transplant (FMT) transfer efficiency is an underappreciated source of experimental variability in human microbiota associated (HMA) mice. Pooled human IBD patient fecal microbiota engrafted germ-free (GF) mice with low amplicon sequence variant (ASV)-level transfer efficiency, resulting in high recipient-to-recipient variation of microbiota composition and colitis severity in HMA Il-10-/- mice. In contrast, mouse-to-mouse transfer of mouse-adapted human IBD patient microbiota transferred with high efficiency and low compositional variability resulting in highly consistent and reproducible colitis phenotypes in recipient Il-10-/- mice. Human-to-mouse FMT caused a population bottleneck with reassembly of microbiota composition that was host inflammatory environment specific. Mouse-adaptation in the inflamed Il-10-/- host reassembled a more aggressive microbiota that induced more severe colitis in serial transplant to Il-10-/- mice than the distinct microbiota reassembled in non-inflamed WT hosts. Our findings support a model of IBD pathogenesis in which host inflammation promotes aggressive resident bacteria, which further drives a feed-forward process of dysbiosis exacerbated gut inflammation. This model implies that effective management of IBD requires treating both the dysregulated host immune response and aggressive inflammation-driven microbiota. We propose that our mouse-adapted human microbiota model is an optimized, reproducible, and rigorous system to study human microbiome-driven disease phenotypes, which may be generalized to mouse models of other human microbiota-modulated diseases, including metabolic syndrome/obesity, diabetes, autoimmune diseases, and cancer.
Keywords: Inflammatory bowel diseases; experimental colitis; fecal microbiota transplant; human microbiota associated mice; interleukin-10 deficient; microbiota transfer efficiency; mouse-adapted.
Conflict of interest statement
Competing Interests None relevant to this study. RBS receives grant support from Gusto Global LLC, Biomica, and ImmunyX, and serves on the Scientific Advisory Board of Biomica.
Figures

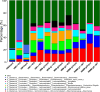

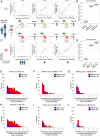

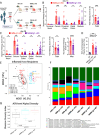
References
-
- Alatab S, Sepanlou SG, Ikuta K, Vahedi H, Bisignano C, Safiri S, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Gastroenterology & Hepatology. 2020;5(1):17–30. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
