This is a preprint.
A Conserved Mechanism of Cardiac Hypertrophy Regression through FoxO1
- PMID: 38328143
- PMCID: PMC10849654
- DOI: 10.1101/2024.01.27.577585
A Conserved Mechanism of Cardiac Hypertrophy Regression through FoxO1
Update in
-
Regression of postprandial cardiac hypertrophy in burmese pythons is mediated by FoxO1.Proc Natl Acad Sci U S A. 2024 Oct 8;121(41):e2408719121. doi: 10.1073/pnas.2408719121. Epub 2024 Oct 1. Proc Natl Acad Sci U S A. 2024. PMID: 39352930 Free PMC article.
Abstract
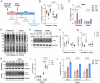
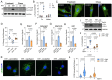
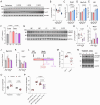
The heart is a highly plastic organ that responds to diverse stimuli to modify form and function. The molecular mechanisms of adaptive physiological cardiac hypertrophy are well-established; however, the regulation of hypertrophy regression is poorly understood. To identify molecular features of regression, we studied Burmese pythons which experience reversible cardiac hypertrophy following large, infrequent meals. Using multi-omics screens followed by targeted analyses, we found forkhead box protein O1 (FoxO1) transcription factor signaling, and downstream autophagy activity, were downregulated during hypertrophy, but re-activated with regression. To determine whether these events were mechanistically related to regression, we established an in vitro platform of cardiomyocyte hypertrophy and regression from treatment with fed python plasma. FoxO1 inhibition prevented regression in this system, while FoxO1 activation reversed fed python plasma-induced hypertrophy in an autophagy-dependent manner. We next examined whether FoxO1 was implicated in mammalian models of reversible hypertrophy from exercise and pregnancy and found that in both cases FoxO1 was activated during regression. In these models, as in pythons, activation of FoxO1 was associated with increased expression FoxO1 target genes involved in autophagy. Taken together, our findings suggest FoxO1-dependent autophagy is a conserved mechanism for regression of physiological cardiac hypertrophy across species.
Conflict of interest statement
COMPETING INTERESTS LAL is a Co-Founder of MyoKardia, acquired by Bristol Myers Squibb. MyoKardia and Bristol Myers Squibb were not involved in this study. The other authors have no competing interests to disclose.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
