This is a preprint.
Widespread Peptide Surfactants with Post-translational C-methylations Promote Bacterial Development
- PMID: 38328144
- PMCID: PMC10849626
- DOI: 10.1101/2024.01.23.576971
Widespread Peptide Surfactants with Post-translational C-methylations Promote Bacterial Development
Update in
-
Peptide surfactants with post-translational C-methylations that promote bacterial development.Nat Chem Biol. 2025 Jul;21(7):1069-1075. doi: 10.1038/s41589-025-01882-8. Epub 2025 Apr 22. Nat Chem Biol. 2025. PMID: 40263466 Free PMC article.
Abstract
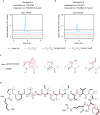
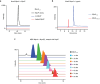
Bacteria produce a variety of peptides to mediate nutrient acquisition, microbial interactions, and other physiological processes. Of special interest are surface-active peptides that aid in growth and development. Herein, we report the structure and characterization of clavusporins, unusual and hydrophobic ribosomal peptides with multiple C-methylations at unactivated carbon centers, which help drastically reduce the surface tension of water and thereby aid in Streptomyces development. The peptides are synthesized by a previously uncharacterized protein superfamily, termed DUF5825, in conjunction with a vitamin B12-dependent radical S-adenosylmethionine metalloenzyme. The operon encoding clavusporin is wide-spread among actinomycete bacteria, suggesting a prevalent role for clavusporins as morphogens in erecting aerial hyphae and thereby advancing sporulation and proliferation.
Conflict of interest statement
Disclosures The authors declare no competing interests.
Figures






Similar articles
-
Peptide surfactants with post-translational C-methylations that promote bacterial development.Nat Chem Biol. 2025 Jul;21(7):1069-1075. doi: 10.1038/s41589-025-01882-8. Epub 2025 Apr 22. Nat Chem Biol. 2025. PMID: 40263466 Free PMC article.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Active body surface warming systems for preventing complications caused by inadvertent perioperative hypothermia in adults.Cochrane Database Syst Rev. 2016 Apr 21;4(4):CD009016. doi: 10.1002/14651858.CD009016.pub2. Cochrane Database Syst Rev. 2016. PMID: 27098439 Free PMC article.
-
Maternal and neonatal outcomes of elective induction of labor.Evid Rep Technol Assess (Full Rep). 2009 Mar;(176):1-257. Evid Rep Technol Assess (Full Rep). 2009. PMID: 19408970 Free PMC article.
-
Smoking cessation medicines and e-cigarettes: a systematic review, network meta-analysis and cost-effectiveness analysis.Health Technol Assess. 2021 Oct;25(59):1-224. doi: 10.3310/hta25590. Health Technol Assess. 2021. PMID: 34668482
References
-
- Clardy J. & Walsh C. Lessons from natural molecules. Nature 432, 829–837 (2004). - PubMed
-
- Wang R. & Seyedsayamdost M. R. Hijacking exogenous signals to generate new secondary metabolites during symbiotic interactions. Nat. Rev. Chem. 1, 0021 (2017).
-
- van der Meij A., Worsley S. F., Hutchings M. I. & van Wezel G. P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416 (2017). - PubMed
-
- Bassler B. L. & Losick R. Bacterially speaking. Cell 125, 237–246 (2006). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
