This is a preprint.
Multi-Channel smFRET study reveals a Compact conformation of EF-G on the Ribosome
- PMID: 38328191
- PMCID: PMC10849647
- DOI: 10.1101/2024.01.27.577133
Multi-Channel smFRET study reveals a Compact conformation of EF-G on the Ribosome
Update in
-
Multi-channel smFRET study reveals a compact conformation of EF-G on the ribosome.Int J Biochem Cell Biol. 2025 Jul;184:106782. doi: 10.1016/j.biocel.2025.106782. Epub 2025 Apr 8. Int J Biochem Cell Biol. 2025. PMID: 40210088 Free PMC article.
Abstract
While elongation factor G (EF-G) is crucial for ribosome translocation, the role of its GTP hydrolysis remains ambiguous. EF-G's indispensability is further exemplified by the phosphorylation of human eukaryotic elongation factor 2 (eEF2) at Thr56, which inhibits protein synthesis globally, but its exact mechanism is not clear. In this study, we developed a multi-channel single-molecule FRET (smFRET) microscopy methodology to examine the conformational changes of E. coli EF-G induced by mutations that closely aligned with eEF2's Thr56 residue. We utilized Alexa 488/594 double-labeled EF-G to catalyze the translocation of fMet-Phe-tRNAPhe-Cy3 inside Cy5-L27 labeled ribosomes, allowing us to probe both processes within the same complex. Our findings indicate that in the presence of either GTP or GDPCP, wild-type EF-G undergoes a conformational extension upon binding to the ribosome to promote normal translocation. On the other hand, T48E and T48V mutations did not affect GTP/GDP binding or GTP hydrolysis, but impeded Poly(Phe) synthesis and caused EF-G to adopt a unique compact conformation, which wasn't observed when the mutants interact solely with the sarcin/ricin loop. This study provides new insights into EF-G's adaptability and sheds light on the modification mechanism of human eEF2.
Keywords: compact EF-G; induced-fit; multi-channel smFRET; ribosome translocation; single molecule FRET.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures


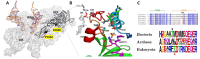

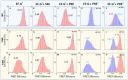
References
-
- Schmeing T. M., Ramakrishnan V., What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 (2009). - PubMed
-
- Connell S. R. et al. , Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell 25, 751–764 (2007). - PubMed
-
- Gonzalez-Garcia J. S., A model for ribosome translocation based on the alternated displacement of its subunits. Eur Biophys J 52, 175–187 (2023). - PubMed
-
- Schuwirth B. S. et al. , Structures of the bacterial ribosome at 3.5 A resolution. Science 310, 827–834 (2005). - PubMed
-
- Frank J., Agrawal R. K., A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322 (2000). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
