This is a preprint.
Patient derived model of UBA5-associated encephalopathy identifies defects in neurodevelopment and highlights potential therapies
- PMID: 38328212
- PMCID: PMC10849720
- DOI: 10.1101/2024.01.25.577254
Patient derived model of UBA5-associated encephalopathy identifies defects in neurodevelopment and highlights potential therapies
Update in
-
Patient-derived models of UBA5-associated encephalopathy identify defects in neurodevelopment and highlight potential therapeutic avenues.Sci Transl Med. 2025 May 7;17(797):eadn8417. doi: 10.1126/scitranslmed.adn8417. Epub 2025 May 7. Sci Transl Med. 2025. PMID: 40333994
Abstract
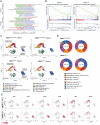
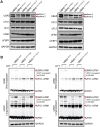
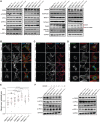
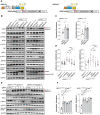
UBA5 encodes for the E1 enzyme of the UFMylation cascade, which plays an essential role in ER homeostasis. The clinical phenotypes of UBA5-associated encephalopathy include developmental delays, epilepsy and intellectual disability. To date, there is no humanized neuronal model to study the cellular and molecular consequences of UBA5 pathogenic variants. We developed and characterized patient-derived cortical organoid cultures and identified defects in GABAergic interneuron development. We demonstrated aberrant neuronal firing and microcephaly phenotypes in patient-derived organoids. Mechanistically, we show that ER homeostasis is perturbed along with exacerbated unfolded protein response pathway in cells and organoids expressing UBA5 pathogenic variants. We also assessed two gene expression modalities that augmented UBA5 expression to rescue aberrant molecular and cellular phenotypes. Our study provides a novel humanized model that allows further investigations of UBA5 variants in the brain and highlights novel systemic approaches to alleviate cellular aberrations for this rare, developmental disorder.
Conflict of interest statement
Competing interests: The authors have declared that no conflict of interest exists.
Figures




References
-
- Mignon-Ravix C. et al., Abnormal function of the UBA5 protein in a case of early developmental and epileptic encephalopathy with suppression-burst. Hum Mutat 39, 934–938 (2018). - PubMed
-
- Low K. J. et al., Hemizygous UBA5 missense mutation unmasks recessive disorder in a patient with infantile-onset encephalopathy, acquired microcephaly, small cerebellum, movement disorder and severe neurodevelopmental delay. Eur J Med Genet 62, 97–102 (2019). - PubMed
-
- Daida A. et al., Biallelic loss-of-function UBA5 mutations in a patient with intractable West syndrome and profound failure to thrive. Epileptic Disord 20, 313–318 (2018). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
