This is a preprint.
Identification of Family-Specific Features in Cas9 and Cas12 Proteins: A Machine Learning Approach Using Complete Protein Feature Spectrum
- PMID: 38328240
- PMCID: PMC10849529
- DOI: 10.1101/2024.01.22.576286
Identification of Family-Specific Features in Cas9 and Cas12 Proteins: A Machine Learning Approach Using Complete Protein Feature Spectrum
Update in
-
Identification of Family-Specific Features in Cas9 and Cas12 Proteins: A Machine Learning Approach Using Complete Protein Feature Spectrum.J Chem Inf Model. 2024 Jun 24;64(12):4897-4911. doi: 10.1021/acs.jcim.4c00625. Epub 2024 Jun 5. J Chem Inf Model. 2024. PMID: 38838358
Abstract
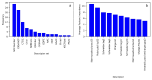
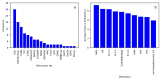
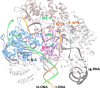
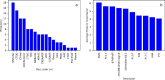
The recent development of CRISPR-Cas technology holds promise to correct gene-level defects for genetic diseases. The key element of the CRISPR-Cas system is the Cas protein, a nuclease that can edit the gene of interest assisted by guide RNA. However, these Cas proteins suffer from inherent limitations like large size, low cleavage efficiency, and off-target effects, hindering their widespread application as a gene editing tool. Therefore, there is a need to identify novel Cas proteins with improved editing properties, for which it is necessary to understand the underlying features governing the Cas families. In the current study, we aim to elucidate the unique protein attributes associated with Cas9 and Cas12 families and identify the features that distinguish each family from the other. Here, we built Random Forest (RF) binary classifiers to distinguish Cas12 and Cas9 proteins from non-Cas proteins, respectively, using the complete protein feature spectrum (13,495 features) encoding various physiochemical, topological, constitutional, and coevolutionary information of Cas proteins. Furthermore, we built multiclass RF classifiers differentiating Cas9, Cas12, and Non-Cas proteins. All the models were evaluated rigorously on the test and independent datasets. The Cas12 and Cas9 binary models achieved a high overall accuracy of 95% and 97% on their respective independent datasets, while the multiclass classifier achieved a high F1 score of 0.97. We observed that Quasi-sequence-order descriptors like Schneider-lag descriptors and Composition descriptors like charge, volume, and polarizability are essential for the Cas12 family. More interestingly, we discovered that Amino Acid Composition descriptors, especially the Tripeptide Composition (TPC) descriptors, are important for the Cas9 family. Four of the identified important descriptors of Cas9 classification are tripeptides PWN, PYY, HHA, and DHI, which are seen to be conserved across all the Cas9 proteins and were located within different catalytically important domains of the Cas9 protein structure. Among these four tripeptides, tripeptides DHI and HHA are well-known to be involved in the DNA cleavage activity of the Cas9 protein. We therefore propose the the other two tripeptides, PWN and PYY, may also be essential for the Cas9 family. Our identified important descriptors enhanced the understanding of the catalytic mechanisms of Cas9 and Cas12 proteins and provide valuable insights into design of novel Cas systems to achieve enhanced gene-editing properties.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Rath D., Amlinger L., Rath A. & Lundgren M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128 - PubMed
-
- Barrangou R. et al. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. - PubMed
-
- Bolotin A., Quinquis B., Sorokin A. & Ehrlich S. D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
