Secreted Protein Profiling of Human Aortic Smooth Muscle Cells Identifies Vascular Disease Associations
- PMID: 38328934
- PMCID: PMC10978267
- DOI: 10.1161/ATVBAHA.123.320274
Secreted Protein Profiling of Human Aortic Smooth Muscle Cells Identifies Vascular Disease Associations
Abstract
Background: Smooth muscle cells (SMCs), which make up the medial layer of arteries, are key cell types involved in cardiovascular disease, the leading cause of mortality and morbidity worldwide. In response to microenvironment alterations, SMCs dedifferentiate from a contractile to a synthetic phenotype characterized by an increased proliferation, migration, production of ECM (extracellular matrix) components, and decreased expression of SMC-specific contractile markers. These phenotypic changes result in vascular remodeling and contribute to the pathogenesis of cardiovascular disease, including coronary artery disease, stroke, hypertension, and aortic aneurysms. Here, we aim to identify the genetic variants that regulate ECM secretion in SMCs and predict the causal proteins associated with vascular disease-related loci identified in genome-wide association studies.
Methods: Using human aortic SMCs from 123 multiancestry healthy heart transplant donors, we collected the serum-free media in which the cells were cultured for 24 hours and conducted liquid chromatography-tandem mass spectrometry-based proteomic analysis of the conditioned media.
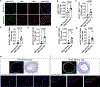
Results: We measured the abundance of 270 ECM and related proteins. Next, we performed protein quantitative trait locus mapping and identified 20 loci associated with secreted protein abundance in SMCs. We functionally annotated these loci using a colocalization approach. This approach prioritized the genetic variant rs6739323-A at the 2p22.3 locus, which is associated with lower expression of LTBP1 (latent-transforming growth factor beta-binding protein 1) in SMCs and atherosclerosis-prone areas of the aorta, and increased risk for SMC calcification. We found that LTBP1 expression is abundant in SMCs, and its expression at mRNA and protein levels was reduced in unstable and advanced atherosclerotic plaque lesions.
Conclusions: Our results unravel the SMC proteome signature associated with vascular disorders, which may help identify potential therapeutic targets to accelerate the pathway to translation.
Keywords: GWAS; cell proliferation; pQTLs; proteomics; smooth muscle cells; vascular remodeling.
Conflict of interest statement
Figures
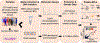

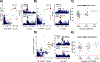

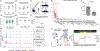

Update of
-
Secreted protein profiling of human aortic smooth muscle cells identifies vascular disease associations.medRxiv [Preprint]. 2023 Nov 10:2023.11.10.23298351. doi: 10.1101/2023.11.10.23298351. medRxiv. 2023. Update in: Arterioscler Thromb Vasc Biol. 2024 Apr;44(4):898-914. doi: 10.1161/ATVBAHA.123.320274. PMID: 37986932 Free PMC article. Updated. Preprint.
References
-
- Alencar GF, Owsiany KM, Karnewar S, Sukhavasi K, Mocci G, Nguyen AT, Williams CM, Shamsuzzaman S, Mokry M, Henderson CA, et al. Stem Cell Pluripotency Genes Klf4 and Oct4 Regulate Complex SMC Phenotypic Changes Critical in Late-Stage Atherosclerotic Lesion Pathogenesis. Circulation 2020;142:2045–2059. doi: 10.1161/CIRCULATIONAHA.120.046672 - DOI - PMC - PubMed
-
- Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, et al. Single-Cell Genomics Reveals a Novel Cell State During Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human. Circulation 2020;142:2060–2075. doi: 10.1161/CIRCULATIONAHA.120.048378 - DOI - PMC - PubMed
-
- Sobue K, Hayashi K, Nishida W. Expressional regulation of smooth muscle cell-specific genes in association with phenotypic modulation. Mol Cell Biochem 1999;190:105–118. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

