Multiscale networks in multiple sclerosis
- PMID: 38329927
- PMCID: PMC10852301
- DOI: 10.1371/journal.pcbi.1010980
Multiscale networks in multiple sclerosis
Abstract
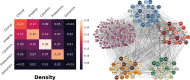
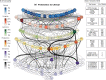
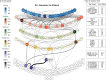
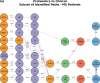
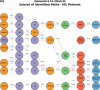
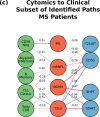
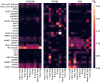
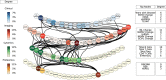
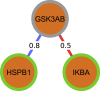
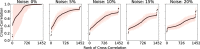
Complex diseases such as Multiple Sclerosis (MS) cover a wide range of biological scales, from genes and proteins to cells and tissues, up to the full organism. In fact, any phenotype for an organism is dictated by the interplay among these scales. We conducted a multilayer network analysis and deep phenotyping with multi-omics data (genomics, phosphoproteomics and cytomics), brain and retinal imaging, and clinical data, obtained from a multicenter prospective cohort of 328 patients and 90 healthy controls. Multilayer networks were constructed using mutual information for topological analysis, and Boolean simulations were constructed using Pearson correlation to identified paths within and among all layers. The path more commonly found from the Boolean simulations connects protein MK03, with total T cells, the thickness of the retinal nerve fiber layer (RNFL), and the walking speed. This path contains nodes involved in protein phosphorylation, glial cell differentiation, and regulation of stress-activated MAPK cascade, among others. Specific paths identified were subsequently analyzed by flow cytometry at the single-cell level. Combinations of several proteins (GSK3AB, HSBP1 or RS6) and immune cells (Th17, Th1 non-classic, CD8, CD8 Treg, CD56 neg, and B memory) were part of the paths explaining the clinical phenotype. The advantage of the path identified from the Boolean simulations is that it connects information about these known biological pathways with the layers at higher scales (retina damage and disability). Overall, the identified paths provide a means to connect the molecular aspects of MS with the overall phenotype.
Copyright: © 2024 Kennedy et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
We have read the journal’s policy and the authors of this manuscript have the following competing interests: KK reports no disclosures. NKdR reports no disclosures. AU received grants and contracts from FISM, Novartis, Biogen, Merck, Fondazione Cariplo, Italian Ministry of Health, received honoraria, or consultation fees from Biogen, Roche, Teva, Merck, Genzyme, Novartis. FI reports no disclosures. MC reports no disclosures. HFH has received honoraria for lecturing or advice from Biogen, Merck, Roche, Novartis and Sanofi. TB has received unrestricted research grants from Biogen and Sanofi-Genzyme. SDB reports no disclosures. EH received honoraria for lecturing and advisory board activity from Biogen, Merck and Sanofi-Genzyme and unrestricted research grant from Merck. SBI reports no disclosures. SAdRB reports no disclosures. FP received honoraria and research support from Alexion, Bayer, Biogen, Chugai, Merck Serono, Novartis, Genzyme, MedImmune, Shire, Teva, and serves on scientific advisory boards for Alexion, MedImmune, and Novartis. He has received funding from Deutsche Forschungsgemeinschaft (DFG Exc 257), Bundesministerium fu?r Bildung und Forschung (Competence Network Multiple Sclerosis), Guthy Jackson Charitable Foundation, EU Framework Program 7, National Multiple Sclerosis Society of the USA. AUB is named as inventor on multiple patents and patents pending owned by Charité - Universitätsmedizin Berlin and/or University of California Irvine for visual computing-based motor function analysis, multiple sclerosis serum biomarkers, and retinal image analysis. He is cofounder and holds shares of Motognosis GmbH and Nocturne GmbH. He serves on the executive board and is Treasurer/Secretary of IMSVISUAL. He received research support from BMWi, BMBF, NIH ICTS, the Kathleen C. Moore Foundation and the Guthy- Jackson Charitable Foundation. Priscilla Ba?cker-Koduah is funded by the DFG Excellence grant to FP (DFG exc 257) and is a Junior scholar of the Einstein Foundation. CC received honoraria for speaking from Bayer and research funding from Novartis, unrelated to this study. SA received a conference grant from Celgene and honoraria for speaking from Alexion, Bayer and Roche. JB reports no disclosures. JSR declares funding from GSK & Sanofi and fees from Travere Therapeutics & Singularity Bio. MR reports no disclosures. LGA is founder and hold stocks at ProtATonce. MA is an employee of Hoffman-La Roche AG, yet this article is related to his activity at the Hospital Clinic of Barcelona. EHML is an employee of the European Medicines Agency (Human Medicines) since 16 April 2019, yet this article is related to her activity at the Hospital Clinic of Barcelona and consequently, it does not in any way represent the views of the Agency or its Committees. SL received compensation for consulting services and speaker honoraria from Biogen Idec, Novartis, TEVA, Genzyme, Sanofi and Merck. AS received compensation for consulting services and speaker honoraria from Bayer-Schering, Merck- Serono, Biogen-Idec, Sanofi-Aventis, TEVA, Novartis and Roche. EMH reports no disclosures. Elisabeth Solana received travel reimbursement from Sanofi and ECTRIMS and reports personal fees from Roche Spain. IPV is currently an employee of UCB pharma, yet this article is related to her activity at the Hospital Clinic of Barcelona. She has received travel reimbursement from Roche Spain and Genzyme-Sanofi, European Academy of Neurology, and European Committee for Treatment and Research in Multiple Sclerosis for international and national meetings over the last 3 years; she holds a patent for an affordable eye-tracking system to measure eye movement in neurologic diseases, and she holds stock in Aura Innovative Robotics. JGO reports no disclosures. PV has received consultancy fees and held stocks from Accure Therapeutics SL, Attune Neurosciences Inc, Spiral Therapeutics Inc, QMenta Inc, CLight Inc, NeuroPrex Inc, StimuSIL and Adhera Health Inc
Figures




References
-
- Ogino S, Jhun I, Mata DA, Soong TR, Hamada T, Liu L, et al. Integration of pharmacology, molecular pathology, and population data science to support precision gastrointestinal oncology. NPJ Precis Oncol. 2017;1:40–8. Epub 2017/01/01. doi: 10.1038/s41698-017-0042-x. ; PubMed Central PMCID: PMC5856171. - DOI - PMC - PubMed
-
- Aleta A, Moreno Y. Multilayer Networks in a Nutshell. Annual Review of Condensed Matter Physics. 2019;10(1):45–62. doi: 10.1146/annurev-conmatphys-031218-013259 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

