Methylthioadenosine Phosphorylase Genomic Loss in Advanced Gastrointestinal Cancers
- PMID: 38330461
- PMCID: PMC11144995
- DOI: 10.1093/oncolo/oyae011
Methylthioadenosine Phosphorylase Genomic Loss in Advanced Gastrointestinal Cancers
Abstract
Background: One of the most common sporadic homozygous deletions in cancers is 9p21 loss, which includes the genes methylthioadenosine phosphorylase (MTAP), CDKN2A, and CDKN2B, and has been correlated with worsened outcomes and immunotherapy resistance. MTAP-loss is a developing drug target through synthetic lethality with MAT2A and PMRT5 inhibitors. The purpose of this study is to investigate the prevalence and genomic landscape of MTAP-loss in advanced gastrointestinal (GI) tumors and investigate its role as a prognostic biomarker.
Materials and methods: We performed next-generation sequencing and comparative genomic and clinical analysis on an extensive cohort of 64 860 tumors comprising 5 GI cancers. We compared the clinical outcomes of patients with GI cancer harboring MTAP-loss and MTAP-intact tumors in a retrospective study.
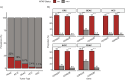
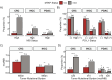
Results: The prevalence of MTAP-loss in GI cancers is 8.30%. MTAP-loss was most prevalent in pancreatic ductal adenocarcinoma (PDAC) at 21.7% and least in colorectal carcinoma (CRC) at 1.1%. MTAP-loss tumors were more prevalent in East Asian patients with PDAC (4.4% vs 3.2%, P = .005) or intrahepatic cholangiocarcinoma (IHCC; 6.4% vs 4.3%, P = .036). Significant differences in the prevalence of potentially targetable genomic alterations (ATM, BRAF, BRCA2, ERBB2, IDH1, PIK3CA, and PTEN) were observed in MTAP-loss tumors and varied according to tumor type. MTAP-loss PDAC, IHCC, and CRC had a lower prevalence of microsatellite instability or elevated tumor mutational burden. Positive PD-L1 tumor cell expression was less frequent among MTAP-loss versus MTAP-intact IHCC tumors (23.2% vs 31.2%, P = .017).
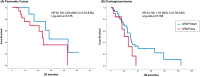
Conclusion: In GI cancers, MTAP-loss occurs as part of 9p21 loss and has an overall prevalence of 8%. MTAP-loss occurs in 22% of PDAC, 15% of IHCC, 8.7% of gastroesophageal adenocarcinoma, 2.4% of hepatocellular carcinoma, and 1.1% of CRC and is not mutually exclusive with other targetable mutations.
Keywords: 9p21 loss; MTAP loss; biomarkers; cholangiocarcinoma; genomics; tumor.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
Dean C. Pavlick, Vamsi Parimi, Richard S.P. Huang, Tyler Janovitz, Natalie Danziger, Mia A. Levy, and Jeffrey S. Ross are current or former employees of Foundation Medicine, a wholly owned subsidiary of Roche, with stock options. Shubham Pant reported consulting or advisory role with Zymeworks, Ipsen, Novartis, Janssen, AskGene Pharma, BPGBio, Jazz, AstraZeneca, Boehringer Ingelheim, USWorldmeds, Nihon Medi-Physics Co, Ltd, and Alligator Bioscience; and research funding (to institution) from Mirati Therapeutics, Lilly, Xencor, Novartis, Bristol-Myers Squibb, Astellas, Framewave, 4D Pharma, Boehringer Ingelheim, NGM Pharmaceuticals, Janssen, Arcus, Elicio, Biontech, Ipsen, Zymeworks, Pfizer, ImmunoMET, Immuneering, and Amal Therapeutics. Milind Javle reported advisory board role or research funding from Abbvie, Array, Astellas, AstraZeneca, Bayer, Beigene, Biocartis, Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Daiichi, GSK, Halozyme, Helsinn, Incyte, Ipsen, Janssen Research, Lilly, Merck Sharp & Dohme, EMD Serono, Novartis, Transthera, Meclun, Eli Lilly, Oncosil, QED, Taiho, Servier, and Agios. The other authors indicated no financial relationships.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

