Bacterial Skin Dysbiosis in Darier Disease
- PMID: 38330926
- PMCID: PMC11168447
- DOI: 10.1159/000537714
Bacterial Skin Dysbiosis in Darier Disease
Abstract
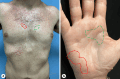
Introduction: Darier disease is a rare inherited disease with dominant skin manifestations including keratotic papules and plaques on sebaceous and flexural areas. Secondary infection of skin lesions is common, and Staphylococcus aureus commonly colonizes these lesions. The aim of the study was to characterize the bacterial microbiome of cutaneous Darier lesions compared to normal-looking skin and disease severity.
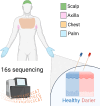
Methods: All patients with a history of Darier followed up at Emek Medical Center were invited to participate in the study. Patients that did not use antibiotics in the past month and signed informed consent had four skin sites sampled with swabs: scalp, chest, axilla, and palm. All samples were analyzed for bacterial microbiome using 16S rDNA sequencing.
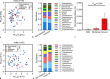
Results: Two hundred and eighty microbiome samples obtained from lesional and non-lesional skin of the scalp, chest, axilla, and palm of 42 Darier patients were included in the analysis. The most abundant bacterial genera across all skin sites were Propionibacterium, Corynebacterium, Paracoccus, Micrococcus, and Anaerococcus. Scalp and chest lesions featured a distinct microbiome configuration that was mainly driven by an overabundance of Staphylococci species. Patients with more severe disease exhibited microbiome alterations in the chest, axilla, and palm compared with patients with only mild disease, driven by Peptoniphilus and Moryella genera in scalp and palmar lesions, respectively.
Conclusion: Staphylococci were significantly associated with Darier lesions and drove Darier-associated dysbiosis. Severity of the disease was associated with two other bacterial genera. Whether these associations also hold a causative role and may serve as a therapeutic target remains to be determined and requires further investigation.
Keywords: Darier disease; Microbiome; Moryella; Peptoniphilus; Staphylococcus.
© 2024 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
The authors state that there is no conflict of interest.
Figures



References
-
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Publ Gr. 2018;16(3):143–55. - PubMed
-
- Brandwein M, Fuks G, Israel A, Sabbah F, Hodak E, Szitenberg A, et al. . Skin microbiome compositional changes in atopic dermatitis accompany dead sea climatotherapy. Photochem Photobiol. 2019;95(6):1446–53. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

