Optimized bacterial absolute quantification method by qPCR using an exogenous bacterial culture as a normalization strategy in triple-species BV-like biofilms
- PMID: 38331102
- PMCID: PMC11149788
- DOI: 10.1016/j.mimet.2024.106895
Optimized bacterial absolute quantification method by qPCR using an exogenous bacterial culture as a normalization strategy in triple-species BV-like biofilms
Abstract
Quantitative Polymerase Chain Reaction (qPCR) is a widely used method in molecular biology to quantify target DNA sequences. Despite its accuracy, there are important experimental controls that should be considered to avoid biased results. One of them is gDNA loss during extraction, which is higher among samples with lower bacterial concentrations. Improvement in qPCR quantification procedures is mandatory to obtain reproducible and accurate results. Herein, we report an improved qPCR method for bacterial quantification of Gardnerella vaginalis, Prevotella bivia, and Fannyhessea vaginae, three key-bacterial vaginosis (BV)-associated bacteria (BVAB) thought to play important roles in the pathogenesis of this common vaginal infection. The formation of a mature biofilm on vaginal epithelial cells is an unique feature of BV and, despite over 60 years of research, the exact etiology of BV remains unknown. Here, we optimized a qPCR method that accurately quantified triple-species biofilms containing these key BVAB, after the addition of an exogenous bacterial control containing a fixed concentration of Escherichia coli, prior to gDNA extraction. This improved method minimized and normalized the inherent losses associated with bacterial centrifugation, which allows better sensitivity at lower bacterial concentrations.
Keywords: Bacterial quantification; Bacterial vaginosis; Calibration curves; Centrifugation loss; Exogenous control; qPCR.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the contents of this manuscript.
Figures
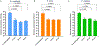


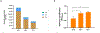
References
-
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, & Wittwer CT (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622. 10.1373/clinchem.2008.112797 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

