ADAM19 cleaves the PTH receptor and associates with brachydactyly type E
- PMID: 38331475
- PMCID: PMC10853454
- DOI: 10.26508/lsa.202302400
ADAM19 cleaves the PTH receptor and associates with brachydactyly type E
Abstract
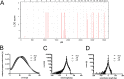
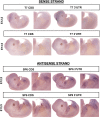
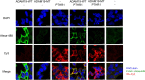
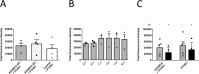
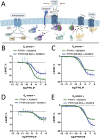
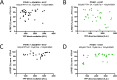
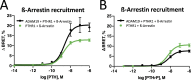
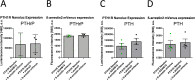
Brachydactyly type E (BDE), shortened metacarpals, metatarsals, cone-shaped epiphyses, and short stature commonly occurs as a sole phenotype. Parathyroid hormone-like protein (PTHrP) has been shown to be responsible in all forms to date, either directly or indirectly. We used linkage and then whole genome sequencing in a small pedigree, to elucidate BDE and identified a truncated disintegrin-and-metalloproteinase-19 (ADAM19) allele in all affected family members, but not in nonaffected persons. Since we had shown earlier that the extracellular domain of the parathyroid hormone receptor (PTHR1) is subject to an unidentified metalloproteinase cleavage, we tested the hypothesis that ADAM19 is a sheddase for PTHR1. WT ADAM19 cleaved PTHR1, while mutated ADAM-19 did not. We mapped the cleavage site that we verified with mass spectrometry between amino acids 64-65. ADAM-19 cleavage increased Gq and decreased Gs activation. Moreover, perturbed PTHR1 cleavage by ADAM19 increased ß-arrestin2 recruitment, while cAMP accumulation was not altered. We suggest that ADAM19 serves as a regulatory element for PTHR1 and could be responsible for BDE. This sheddase may affect other PTHrP or PTH-related functions.
© 2024 Aydin et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures












References
-
- Cartwright JD, Rosin M, Robertson C (1980) Brachydactyly type E: A report of a family. S Afr Med J 58: 255–257. - PubMed
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
