Prediction of ineffectiveness of biological drugs using machine learning and explainable AI methods: data from the Austrian Biological Registry BioReg
- PMID: 38331930
- PMCID: PMC10851538
- DOI: 10.1186/s13075-024-03277-x
Prediction of ineffectiveness of biological drugs using machine learning and explainable AI methods: data from the Austrian Biological Registry BioReg
Abstract
Objectives: Machine learning models can support an individualized approach in the choice of bDMARDs. We developed prediction models for 5 different bDMARDs using machine learning methods based on patient data derived from the Austrian Biologics Registry (BioReg).
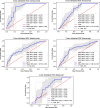
Methods: Data from 1397 patients and 19 variables with at least 100 treat-to-target (t2t) courses per drug were derived from the BioReg biologics registry. Different machine learning algorithms were trained to predict the risk of ineffectiveness for each bDMARD within the first 26 weeks. Cross-validation and hyperparameter optimization were applied to generate the best models. Model quality was assessed by area under the receiver operating characteristic (AUROC). Using explainable AI (XAI), risk-reducing and risk-increasing factors were extracted.
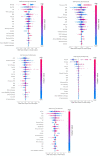
Results: The best models per drug achieved an AUROC score of the following: abatacept, 0.66 (95% CI, 0.54-0.78); adalimumab, 0.70 (95% CI, 0.68-0.74); certolizumab, 0.84 (95% CI, 0.79-0.89); etanercept, 0.68 (95% CI, 0.55-0.87); tocilizumab, 0.72 (95% CI, 0.69-0.77). The most risk-increasing variables were visual analytic scores (VAS) for abatacept and etanercept and co-therapy with glucocorticoids for adalimumab. Dosage was the most important variable for certolizumab and associated with a lower risk of non-response. Some variables, such as gender and rheumatoid factor (RF), showed opposite impacts depending on the bDMARD.
Conclusion: Ineffectiveness of biological drugs could be predicted with promising accuracy. Interestingly, individual parameters were found to be associated with drug responses in different directions, indicating highly complex interactions. Machine learning can be of help in the decision-process by disentangling these relations.
Keywords: DMARDs; Machine learning; Rheumatoid arthritis; Routinely collected data; bDMARD.
© 2024. The Author(s).
Conflict of interest statement
Disclosure of Interests: Dubravka Ukalovic Employee of: Siemens Healthineers, Burkhard Leeb Member of speakers’ bureau: AbbVie, Roche, MSD, Pfizer, Actiopharm, Boehringer-Ingelheim, Kwizda, Celgene, Sandoz, Grünenthal, Eli-Lilly, Consultant of: AbbVie, Amgen, Roche, MSD, Pfizer, Celgene, Grünenthal, Kwizda, Eli-Lilly, Novartis, Sandoz, Received Honoria from: Abbvie, Biogen, Celgene, Eli Lilly, MSD, Pfizer, Roche, Novartis and Sandoz, Bernhard Rintelen Member of speakers’ bureau: BMS, Eli-Lilly, Pfizer, TRB-Chemedica, UCB, Wyeth, Consultant of: Abbott, Abbvie, Amgen, Gileat, Novartis, Pfizer, Roche, TRB-Chemedica, UCB, Wyeth, Grant/research support from: Abbott, Aesca, Amgen, Centocor, Eli-Lilly, Servier, UCB, Gabriela Eichbauer-Sturm Member of speakers’ bureau: AbbVie, Astro-Pharma, Grünenthal, Jansen, Eli-Lilly, Menarini, MSD, Novartis, Pfizer, Roche, TRB, UCB, Fresenius Kabi, Peter Spellitz: None declared, Rudolf Puchner Member of speakers’ bureau: AbbVie, BMS, Janssen, Kwizda, MSD, Pfizer, Celgene, Grünenthal, Eli-Lilly, Consultant of: AbbVie, Amgen, Pfizer, Celgene, Grünenthal, Eli-Lilly, Received Honoria from: Abbvie, BMS, Gilead, Janssen, Kwizda, Lilly, MSD, Novartis and Pfizer, Manfred Herold: None declared, Miriam Stetter: None declared, Vera Ferincz: None declared, Johannes Resch-Passini: None declared, Jochen Zwerina: None declared, Marcus Zimmermann-Rittereiser Employee of: Siemens Healthineers, Shareholder of: Siemens Healthineers, Ruth Fritsch-Stork Member of speakers’ bureau: AbbVie, Astra Zeneca, Astropharm, Novartis.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

