Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor
- PMID: 38332002
- PMCID: PMC10853176
- DOI: 10.1038/s41467-024-45241-y
Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor
Abstract
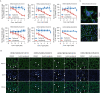
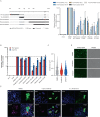
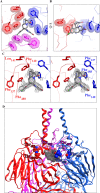
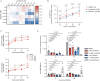
Respiratory syncytial virus (RSV) is a common cause of acute lower respiratory tract infection in infants, older adults and the immunocompromised. Effective directly acting antivirals are not yet available for clinical use. To address this, we screen the ReFRAME drug-repurposing library consisting of 12,000 small molecules against RSV. We identify 21 primary candidates including RSV F and N protein inhibitors, five HSP90 and four IMPDH inhibitors. We select lonafarnib, a licensed farnesyltransferase inhibitor, and phase III candidate for hepatitis delta virus (HDV) therapy, for further follow-up. Dose-response analyses and plaque assays confirm the antiviral activity (IC50: 10-118 nM). Passaging of RSV with lonafarnib selects for phenotypic resistance and fixation of mutations in the RSV fusion protein (T335I and T400A). Lentiviral pseudotypes programmed with variant RSV fusion proteins confirm that lonafarnib inhibits RSV cell entry and that these mutations confer lonafarnib resistance. Surface plasmon resonance reveals RSV fusion protein binding of lonafarnib and co-crystallography identifies the lonafarnib binding site within RSV F. Oral administration of lonafarnib dose-dependently reduces RSV virus load in a murine infection model using female mice. Collectively, this work provides an overview of RSV drug repurposing candidates and establishes lonafarnib as a bona fide fusion protein inhibitor.
© 2024. The Author(s).
Conflict of interest statement
SMS, SH, JR, TFS and TP disclose that they are authoring a patent application “LONAFARNIB FOR USE IN THE TREATMENT OF VIRAL INFECTIONS” describing lonafarnib as antiviral compound that inhibits RSV infection (21152993.8-1132). The authors declare no competing interests and no restrictions on the publication of data.
Figures




References
-
- Haapanen M, Renko M, Artama M, Kuitunen I. The impact of the lockdown and the re-opening of schools and day cares on the epidemiology of SARS-CoV-2 and other respiratory infections in children—A nationwide register study in Finland. EClinicalMedicine. 2021;34:100807. doi: 10.1016/j.eclinm.2021.100807. - DOI - PMC - PubMed
-
- Bardsley M, et al. Epidemiology of respiratory syncytial virus in children younger than 5 years in England during the COVID-19 pandemic, measured by laboratory, clinical, and syndromic surveillance: a retrospective observational study. Lancet Infect Dis. 2023;23:56–66. doi: 10.1016/S1473-3099(22)00525-4. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- EXC 2155 RESIST, Project 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- EXC 2155 "RESIST" - Project ID 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- EXC 2155 "RESIST" - Project ID 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- EXC 2155 "RESIST" - Project ID 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- EXC 2155 "RESIST" - Project ID 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- EXC2155 RESIST_390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- INDIRA 11-76251-99-6/19 (ZN3437)/Niedersächsisches Ministerium für Wissenschaft und Kultur (Ministry for Science and Culture of Lower Saxony)
- OPTIS, Project ID 9B811/Volkswagen Foundation (VolkswagenStiftung)
- OPTIS, Project ID 9B811/Volkswagen Foundation (VolkswagenStiftung)
- OPTIS, Project ID 9B811/Volkswagen Foundation (VolkswagenStiftung)
- OPTIS, ID9B811/Volkswagen Foundation (VolkswagenStiftung)
- DZIF, TTU 09.719/Deutsches Zentrum für Infektionsforschung (German Center for Infection Research)
- INDIRA 11-76251-99-6/19 (ZN3437)/Niedersächsische Ministerium für Wissenschaft und Kultur (Lower Saxony Ministry of Science and Culture)
LinkOut - more resources
Full Text Sources
Medical

