ARL5b inhibits human rhinovirus 16 propagation and impairs macrophage-mediated bacterial clearance
- PMID: 38332148
- PMCID: PMC10933434
- DOI: 10.1038/s44319-024-00069-x
ARL5b inhibits human rhinovirus 16 propagation and impairs macrophage-mediated bacterial clearance
Abstract
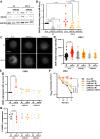
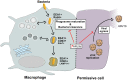
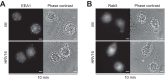
Human rhinovirus is the most frequently isolated virus during severe exacerbations of chronic respiratory diseases, like chronic obstructive pulmonary disease. In this disease, alveolar macrophages display significantly diminished phagocytic functions that could be associated with bacterial superinfections. However, how human rhinovirus affects the functions of macrophages is largely unknown. Macrophages treated with HRV16 demonstrate deficient bacteria-killing activity, impaired phagolysosome biogenesis, and altered intracellular compartments. Using RNA sequencing, we identify the small GTPase ARL5b to be upregulated by the virus in primary human macrophages. Importantly, depletion of ARL5b rescues bacterial clearance and localization of endosomal markers in macrophages upon HRV16 exposure. In permissive cells, depletion of ARL5b increases the secretion of HRV16 virions. Thus, we identify ARL5b as a novel regulator of intracellular trafficking dynamics and phagolysosomal biogenesis in macrophages and as a restriction factor of HRV16 in permissive cells.
Keywords: Bacteria; Endosomes; Macrophages; Phagosome Maturation; Restriction Factor.
© 2024. The Author(s).
Conflict of interest statement
LO, EI, KT, GM, DC, and NK work or worked for AstraZeneca which develops treatments and vaccines against pathogens. The remaining authors declare no competing interests.
Figures






References
-
- Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, Korlach J, Bakaletz LO, Jennings MP, Peak IR, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014;28:5197–5207. doi: 10.1096/fj.14-256578. - DOI - PubMed
MeSH terms
Grants and funding
- ARF202110013926/Fondation pour la Recherche Médicale (FRM)
- MacroRhino/EC | Horizon Europe | Excellent Science | HORIZON EUROPE Marie Sklodowska-Curie Actions (MSCA)
- ANR-10-INSB-04/Agence Nationale de la Recherche (ANR)
- ANR-10-INBS-04/Agence Nationale de la Recherche (ANR)
- ANR-11-LABX-0038/Labex CelTisPhyBio (Celtisphybio Labex)
LinkOut - more resources
Full Text Sources

