Crosstalk between CD64+MHCII+ macrophages and CD4+ T cells drives joint pathology during chikungunya
- PMID: 38332201
- PMCID: PMC10940729
- DOI: 10.1038/s44321-024-00028-y
Crosstalk between CD64+MHCII+ macrophages and CD4+ T cells drives joint pathology during chikungunya
Abstract
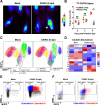
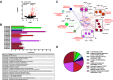
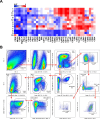
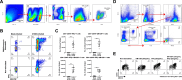
Communications between immune cells are essential to ensure appropriate coordination of their activities. Here, we observed the infiltration of activated macrophages into the joint-footpads of chikungunya virus (CHIKV)-infected animals. Large numbers of CD64+MHCII+ and CD64+MHCII- macrophages were present in the joint-footpad, preceded by the recruitment of their CD11b+Ly6C+ inflammatory monocyte precursors. Recruitment and differentiation of these myeloid subsets were dependent on CD4+ T cells and GM-CSF. Transcriptomic and gene ontology analyses of CD64+MHCII+ and CD64+MHCII- macrophages revealed 89 differentially expressed genes, including genes involved in T cell proliferation and differentiation pathways. Depletion of phagocytes, including CD64+MHCII+ macrophages, from CHIKV-infected mice reduced disease pathology, demonstrating that these cells play a pro-inflammatory role in CHIKV infection. Together, these results highlight the synergistic dynamics of immune cell crosstalk in driving CHIKV immunopathogenesis. This study provides new insights in the disease mechanism and offers opportunities for development of novel anti-CHIKV therapeutics.
Keywords: Chikungunya; GM-CSF; Immune Crosstalk; Immunopathogenesis.
© 2024. The Author(s).
Figures








References
-
- Amorim A, De Feo D, Friebel E, Ingelfinger F, Anderfuhren CD, Krishnarajah S, Andreadou M, Welsh CA, Liu Z, Ginhoux F, et al. IFNgamma and GM-CSF control complementary differentiation programs in the monocyte-to-phagocyte transition during neuroinflammation. Nat Immunol. 2022;23:217–228. - PubMed
-
- Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. - PubMed
-
- Becher B, Tugues S, Greter M. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity. 2016;45:963–973. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

