Human archetypal pluripotent stem cells differentiate into trophoblast stem cells via endogenous BMP5/7 induction without transitioning through naive state
- PMID: 38332235
- PMCID: PMC10853519
- DOI: 10.1038/s41598-024-53381-w
Human archetypal pluripotent stem cells differentiate into trophoblast stem cells via endogenous BMP5/7 induction without transitioning through naive state
Abstract
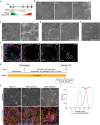
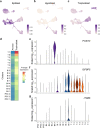
Primary human trophoblast stem cells (TSCs) and TSCs derived from human pluripotent stem cells (hPSCs) can potentially model placental processes in vitro. Yet, the pluripotent states and factors involved in the differentiation of hPSCs to TSCs remain poorly understood. In this study, we demonstrate that the primed pluripotent state can generate TSCs by activating pathways such as Epidermal Growth Factor (EGF) and Wingless-related integration site (WNT), and by suppressing tumor growth factor beta (TGFβ), histone deacetylases (HDAC), and Rho-associated protein kinase (ROCK) signaling pathways, all without the addition of exogenous Bone morphogenetic protein 4 (BMP4)-a condition we refer to as the TS condition. We characterized this process using temporal single-cell RNA sequencing to compare TS conditions with differentiation protocols involving BMP4 activation alone or BMP4 activation in conjunction with WNT inhibition. The TS condition consistently produced a stable, proliferative cell type that closely mimics first-trimester placental cytotrophoblasts, marked by the activation of endogenous retroviral genes and the absence of amnion expression. This was observed across multiple cell lines, including various primed induced pluripotent stem cell (iPSC) and embryonic stem cell (ESC) lines. Primed-derived TSCs can proliferate for over 30 passages and further specify into multinucleated syncytiotrophoblasts and extravillous trophoblast cells. Our research establishes that the differentiation of primed hPSCs to TSC under TS conditions triggers the induction of TMSB4X, BMP5/7, GATA3, and TFAP2A without progressing through a naive state. These findings propose that the primed hPSC state is part of a continuum of potency with the capacity to differentiate into TSCs through multiple routes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

