Mitochondrial DNA replication stress triggers a pro-inflammatory endosomal pathway of nucleoid disposal
- PMID: 38332353
- PMCID: PMC11026068
- DOI: 10.1038/s41556-023-01343-1
Mitochondrial DNA replication stress triggers a pro-inflammatory endosomal pathway of nucleoid disposal
Abstract
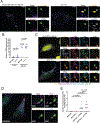
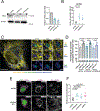
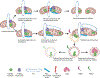
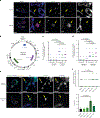
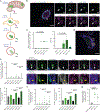
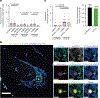
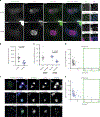
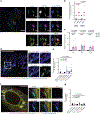
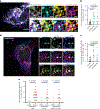
Mitochondrial DNA (mtDNA) encodes essential subunits of the oxidative phosphorylation system, but is also a major damage-associated molecular pattern (DAMP) that engages innate immune sensors when released into the cytoplasm, outside of cells or into circulation. As a DAMP, mtDNA not only contributes to anti-viral resistance, but also causes pathogenic inflammation in many disease contexts. Cells experiencing mtDNA stress caused by depletion of the mtDNA-packaging protein, transcription factor A, mitochondrial (TFAM) or during herpes simplex virus-1 infection exhibit elongated mitochondria, enlargement of nucleoids (mtDNA-protein complexes) and activation of cGAS-STING innate immune signalling via mtDNA released into the cytoplasm. However, the relationship among aberrant mitochondria and nucleoid dynamics, mtDNA release and cGAS-STING activation remains unclear. Here we show that, under a variety of mtDNA replication stress conditions and during herpes simplex virus-1 infection, enlarged nucleoids that remain bound to TFAM exit mitochondria. Enlarged nucleoids arise from mtDNA experiencing replication stress, which causes nucleoid clustering via a block in mitochondrial fission at a stage when endoplasmic reticulum actin polymerization would normally commence, defining a fission checkpoint that ensures mtDNA has completed replication and is competent for segregation into daughter mitochondria. Chronic engagement of this checkpoint results in enlarged nucleoids trafficking into early and then late endosomes for disposal. Endosomal rupture during transit through this endosomal pathway ultimately causes mtDNA-mediated cGAS-STING activation. Thus, we propose that replication-incompetent nucleoids are selectively eliminated by an adaptive mitochondria-endosomal quality control pathway that is prone to innate immune system activation, which might represent a therapeutic target to prevent mtDNA-mediated inflammation during viral infection and other pathogenic states.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

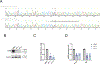
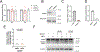






References
-
- Kasashima K, Sumitani M & Endo H Human mitochondrial transcription factor A is required for the segregation of mitochondrial DNA in cultured cells. Exp. Cell Res. 317, 210–220 (2011). - PubMed
-
- Bonawitz ND, Clayton DA & Shadel GS Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell 24, 813–825 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

