The 'analysis of gene expression and biomarkers for point-of-care decision support in Sepsis' study; temporal clinical parameter analysis and validation of early diagnostic biomarker signatures for severe inflammation andsepsis-SIRS discrimination
- PMID: 38332914
- PMCID: PMC10850284
- DOI: 10.3389/fimmu.2023.1308530
The 'analysis of gene expression and biomarkers for point-of-care decision support in Sepsis' study; temporal clinical parameter analysis and validation of early diagnostic biomarker signatures for severe inflammation andsepsis-SIRS discrimination
Abstract
Introduction: Early diagnosis of sepsis and discrimination from SIRS is crucial for clinicians to provide appropriate care, management and treatment to critically ill patients. We describe identification of mRNA biomarkers from peripheral blood leukocytes, able to identify severe, systemic inflammation (irrespective of origin) and differentiate Sepsis from SIRS, in adult patients within a multi-center clinical study.
Methods: Participants were recruited in Intensive Care Units (ICUs) from multiple UK hospitals, including fifty-nine patients with abdominal sepsis, eighty-four patients with pulmonary sepsis, forty-two SIRS patients with Out-of-Hospital Cardiac Arrest (OOHCA), sampled at four time points, in addition to thirty healthy control donors. Multiple clinical parameters were measured, including SOFA score, with many differences observed between SIRS and sepsis groups. Differential gene expression analyses were performed using microarray hybridization and data analyzed using a combination of parametric and non-parametric statistical tools.
Results: Nineteen high-performance, differentially expressed mRNA biomarkers were identified between control and combined SIRS/Sepsis groups (FC>20.0, p<0.05), termed 'indicators of inflammation' (I°I), including CD177, FAM20A and OLAH. Best-performing minimal signatures e.g. FAM20A/OLAH showed good accuracy for determination of severe, systemic inflammation (AUC>0.99). Twenty entities, termed 'SIRS or Sepsis' (S°S) biomarkers, were differentially expressed between sepsis and SIRS (FC>2·0, p-value<0.05).
Discussion: The best performing signature for discriminating sepsis from SIRS was CMTM5/CETP/PLA2G7/MIA/MPP3 (AUC=0.9758). The I°I and S°S signatures performed variably in other independent gene expression datasets, this may be due to technical variation in the study/assay platform.
Keywords: SIRS; biomarker; diagnostic; mRNA signature; sepsis; severe inflammation.
Copyright © 2024 Szakmany, Fitzgerald, Garlant, Whitehouse, Molnar, Shah, Tong, Hall, Ball and Kempsell.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
 , PLMN S
, PLMN S  , SIRS DNS
, SIRS DNS  , SIRS S
, SIRS S  , ABDM DNS
, ABDM DNS  , ABDM S
, ABDM S  .
.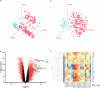
 versus combined SIRS&Sepsis biomarker groups
versus combined SIRS&Sepsis biomarker groups  (each symbol depicting an individual within each group) (B) volcano plot of log10 p-value vs log fold-change of all gene entities, using a 2-fold change cutoff and with select I°I genes highlighted (C) PCA analysis of CNTRL
(each symbol depicting an individual within each group) (B) volcano plot of log10 p-value vs log fold-change of all gene entities, using a 2-fold change cutoff and with select I°I genes highlighted (C) PCA analysis of CNTRL  versus combined SIRS&Sepsis biomarker groups
versus combined SIRS&Sepsis biomarker groups  (each symbol depicting an individual within each group) using select I°I genes only (from
Table 2
) (D) heat map of select I°I biomarkers from
Table 2
across all control, SIRS, ABDM and PLMN sepsis groups stratified by day and prognosis (died/survived).
(each symbol depicting an individual within each group) using select I°I genes only (from
Table 2
) (D) heat map of select I°I biomarkers from
Table 2
across all control, SIRS, ABDM and PLMN sepsis groups stratified by day and prognosis (died/survived).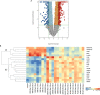
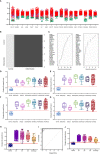
 , SIRS
, SIRS  , Sepsis
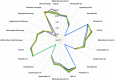
, Sepsis  (B) Random forest classification of validation data into controls and inflammation groups with ‘mtry’ of 31, ‘ntree’ of 2001 (C) Visualization of random forest models features of importance ranked by mean decrease accuracy and mean decrease Gini score (D) I°I candidate panel: ADM+CD177+FAM20A+ITGA7+MMP9+OLAH (E) I°I candidate panel: ADM+FAM20A+ OLAH+ITGA7+MPP9 (F) I°I candidate panel: ADM+OLAH+ FAM20A (G) I°I candidate panel: OLAH+FAM20A (H) I°I candidate panel: ADM+FAM20A+ OLAH+ITGA7+MMP9 across all time-points (I) ROC curves of ADM+FAM20A+OLAH ITGA7+MMP9 and OLAH+FAM20A comparing CNTRL vs SIRS/sepsis across day 1, day 2, day 5 and discharge time points (J) I°I candidate panel: OLAH+FAM20A across all timepoints.
(B) Random forest classification of validation data into controls and inflammation groups with ‘mtry’ of 31, ‘ntree’ of 2001 (C) Visualization of random forest models features of importance ranked by mean decrease accuracy and mean decrease Gini score (D) I°I candidate panel: ADM+CD177+FAM20A+ITGA7+MMP9+OLAH (E) I°I candidate panel: ADM+FAM20A+ OLAH+ITGA7+MPP9 (F) I°I candidate panel: ADM+OLAH+ FAM20A (G) I°I candidate panel: OLAH+FAM20A (H) I°I candidate panel: ADM+FAM20A+ OLAH+ITGA7+MMP9 across all time-points (I) ROC curves of ADM+FAM20A+OLAH ITGA7+MMP9 and OLAH+FAM20A comparing CNTRL vs SIRS/sepsis across day 1, day 2, day 5 and discharge time points (J) I°I candidate panel: OLAH+FAM20A across all timepoints.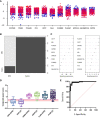
 , ABDM
, ABDM  , PLMN
, PLMN  . (B) Random forest classification of validation data into SIRS and Sepsis ‘mtry’ of 11, ‘ntree’ of 2001 (C) Visualization of random forest models features of importance ranked by mean decrease accuracy and mean decrease Gini score (D) S°S candidate panel: CETP+CMTM5+MIA-MPP3-PLA2G7 (E) ROC curves of CETP+CMTM5+MIA-MPP3-PLA2G7 for SIRS vs Sepsis, SIRS vs Abdominal sepsis and SIRS vs Pulmonary sepsis comparisons.
. (B) Random forest classification of validation data into SIRS and Sepsis ‘mtry’ of 11, ‘ntree’ of 2001 (C) Visualization of random forest models features of importance ranked by mean decrease accuracy and mean decrease Gini score (D) S°S candidate panel: CETP+CMTM5+MIA-MPP3-PLA2G7 (E) ROC curves of CETP+CMTM5+MIA-MPP3-PLA2G7 for SIRS vs Sepsis, SIRS vs Abdominal sepsis and SIRS vs Pulmonary sepsis comparisons.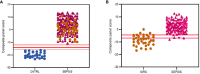
 , SIRS
, SIRS  , SEPSIS
, SEPSIS  (B) the S°S Signature; SIRS
(B) the S°S Signature; SIRS  , PLMN SEPSIS
, PLMN SEPSIS  , ABDM SEPSIS
, ABDM SEPSIS  .
.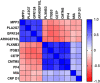
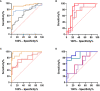
 , healthy controls and non-septic shock
, healthy controls and non-septic shock  , non septic shock and septic shock
, non septic shock and septic shock  (B) GSE9960 comparing healthy controls and sepsis (mixed infection)
(B) GSE9960 comparing healthy controls and sepsis (mixed infection)  , healthy controls and sepsis (gram positive)
, healthy controls and sepsis (gram positive)  , healthy controls and sepsis (gram negative)
, healthy controls and sepsis (gram negative)  healthy controls and sepsis
healthy controls and sepsis  (C) GSE154918 comparing healthy controls and sepsis
(C) GSE154918 comparing healthy controls and sepsis  , healthy controls and follow up of sepsis
, healthy controls and follow up of sepsis  , healthy controls and septic shock
, healthy controls and septic shock  healthy controls and follow up of septic shock
healthy controls and follow up of septic shock  (D) GSE154918 comparing uncomplicated infection and sepsis
(D) GSE154918 comparing uncomplicated infection and sepsis  , uncomplicated infection and follow up of sepsis
, uncomplicated infection and follow up of sepsis  , uncomplicated infection and septic shock
, uncomplicated infection and septic shock  , uncomplicated infection and follow up of septic shock
, uncomplicated infection and follow up of septic shock  .
.References
-
- Daniels R, Nutbeam T. The yellow manual 6th edition. In: The UK sepsis trust. London: United Kingdom Sepsis Trust.
-
- YHEC . The cost of sepsis care in the UK: final report. London: United Kingdom Sepsis Trust. (2017).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

