Whole genome sequencing identified genomic diversity and candidated genes associated with economic traits in Northeasern Merino in China
- PMID: 38333624
- PMCID: PMC10851152
- DOI: 10.3389/fgene.2024.1302222
Whole genome sequencing identified genomic diversity and candidated genes associated with economic traits in Northeasern Merino in China
Abstract
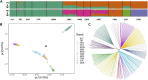
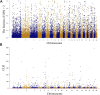
Introduction: Northeast Merino (NMS) is a breed developed in Northeast China during the 1960s for wool and meat production. It exhibits excellent traits such as high wool yield, superior meat quality, rapid growth rate, robust disease resistance, and adaptability to cold climates. However, no studies have used whole-genome sequencing data to investigate the superior traits of NMS. Methods: In this study, we investigated the population structure, genetic diversity, and selection signals of NMS using whole-genome sequencing data from 20 individuals. Two methods (integrated haplotype score and composite likelihood ratio) were used for selection signal analysis, and the Fixation Index was used to explore the selection signals of NMS and the other two breeds, Mongolian sheep and South African meat Merino. Results: The results showed that NMS had low inbreeding levels, high genomic diversity, and a pedigree of both Merino breeds and Chinese local breeds. A total length of 14.09 Mb genomic region containing 287 genes was detected using the two methods. Further exploration of the functions of these genes revealed that they are mainly concentrated in wool production performance (IRF2BP2, MAP3K7, and WNT3), meat production performance (NDUFA9, SETBP1, ZBTB38, and FTO), cold resistance (DNAJC13, LPGAT1, and PRDM16), and immune response (PRDM2, GALNT8, and HCAR2). The selection signals of NMS and the other two breeds annotated 87 and 23 genes, respectively. These genes were also mainly focused on wool and meat production performance. Conclusion: These results provide a basis for further breeding improvement, comprehensive use of this breed, and a reference for research on other breeds.
Keywords: Northeast Merino; genetic diversity; population structure; selection signatures; whole-genome sequencing.
Copyright © 2024 Yi, Hu, Shi, Li, Bai, Sun, Ma, Zhao and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The genomic architecture of South African mutton, pelt, dual-purpose and nondescript sheep breeds relative to global sheep populations.Anim Genet. 2020 Dec;51(6):910-923. doi: 10.1111/age.12991. Epub 2020 Sep 7. Anim Genet. 2020. PMID: 32894610
-
High-resolution analysis of selection sweeps identified between fine-wool Merino and coarse-wool Churra sheep breeds.Genet Sel Evol. 2017 Nov 7;49(1):81. doi: 10.1186/s12711-017-0354-x. Genet Sel Evol. 2017. PMID: 29115919 Free PMC article.
-
Detection of selection signatures in South African Mutton Merino sheep using whole-genome sequencing data.Anim Genet. 2022 Apr;53(2):224-229. doi: 10.1111/age.13173. Epub 2022 Jan 31. Anim Genet. 2022. PMID: 35099062
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
-
Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data.Genes (Basel). 2024 Mar 11;15(3):351. doi: 10.3390/genes15030351. Genes (Basel). 2024. PMID: 38540410 Free PMC article. Review.
Cited by
-
Whole-Genome Resequencing Provides Novel Insights Into the Genetic Diversity, Population Structure, and Patterns of Runs of Homozygosity in Mud Crab (Scylla paramamosain).Evol Appl. 2025 Sep 3;18(9):e70153. doi: 10.1111/eva.70153. eCollection 2025 Sep. Evol Appl. 2025. PMID: 40909995 Free PMC article.
-
Genetic Structure and Selection Signals for Extreme Environment Adaptation in Lop Sheep of Xinjiang.Biology (Basel). 2025 Mar 25;14(4):337. doi: 10.3390/biology14040337. Biology (Basel). 2025. PMID: 40282202 Free PMC article.
-
Relationship between Indel Variants within the JAK2 Gene and Growth Traits in Goats.Animals (Basel). 2024 Jul 6;14(13):1994. doi: 10.3390/ani14131994. Animals (Basel). 2024. PMID: 38998106 Free PMC article.
-
Genomic Inbreeding and Runs of Homozygosity Analysis of Cashmere Goat.Animals (Basel). 2024 Apr 22;14(8):1246. doi: 10.3390/ani14081246. Animals (Basel). 2024. PMID: 38672394 Free PMC article.
References
-
- Banerjee D., Upadhyay R. C., Chaudhary U. B., Kumar R., Singh S., Ashutosh, et al. (2014). Seasonal variation in expression pattern of genes under HSP70: seasonal variation in expression pattern of genes under HSP70 family in heat- and cold-adapted goats (Capra hircus). Cell Stress Chaperones 19 (3), 401–408. 10.1007/s12192-013-0469-0 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

