Culture-independent detection of low-abundant Clostridioides difficile in environmental DNA via PCR
- PMID: 38334406
- PMCID: PMC10952401
- DOI: 10.1128/aem.01278-23
Culture-independent detection of low-abundant Clostridioides difficile in environmental DNA via PCR
Abstract
Clostridioides difficile represents a major burden to public health. As a well-known nosocomial pathogen whose occurrence is highly associated with antibiotic treatment, most examined C. difficile strains originated from clinical specimen and were isolated under selective conditions employing antibiotics. This suggests a significant bias among analyzed C. difficile strains, which impedes a holistic view on this pathogen. In order to support extensive isolation of C. difficile strains from environmental samples, we designed a detection PCR that targets the hpdBCA-operon and thereby identifies low abundances of C. difficile in environmental samples. This operon encodes the 4-hydroxyphenylacetate decarboxylase, which catalyzes the production of the antimicrobial compound para-cresol. Amplicon-based analyses of diverse environmental samples demonstrated that the designed PCR is highly specific for C. difficile and successfully detected C. difficile despite its absence in general 16S rRNA gene-based detection strategies. Further analyses revealed the potential of the hpdBCA detection PCR sequence for initial phylogenetic classification, which allows assessment of C. difficile diversity in environmental samples via amplicon sequencing. Our findings furthermore showed that C. difficile strains isolated under antibiotic treatment from environmental samples were originally dominated by other strains according to PCR amplicon results. This provided evidence for selective cultivation of under-represented but antibiotic-resistant isolates. Thereby, we revealed a substantial bias in C. difficile isolation and research.IMPORTANCEClostridioides difficile is a main cause of diarrheic infections after antibiotic treatment with serious morbidity and mortality worldwide. Research on this pathogen and its virulence has focused on bacterial isolation from clinical specimens under antibiotic treatment, which implies a substantial bias in isolated strains. Comprehensive studies, however, require an unbiased strain collection, which is accomplished by isolation of C. difficile from diverse environmental samples and avoidance of antibiotic-based enrichment strategies. Thus, isolation can significantly benefit from our C. difficile-specific detection PCR, which rapidly verifies C. difficile presence in environmental samples and further allows estimation of the C. difficile diversity by using next-generation sequencing.
Keywords: Clostridioides difficile; antibiotic resistant isolates; detection PCR; environmental DNA; hpdBCA operon; low-abundant microbes; nosocomial infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
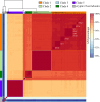


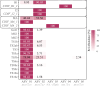
Similar articles
-
Clostridioides difficile para-Cresol Production Is Induced by the Precursor para-Hydroxyphenylacetate.J Bacteriol. 2020 Aug 25;202(18):e00282-20. doi: 10.1128/JB.00282-20. Print 2020 Aug 25. J Bacteriol. 2020. PMID: 32631945 Free PMC article.
-
Regulation of para-cresol production in Clostridioides difficile.Curr Opin Microbiol. 2022 Feb;65:131-137. doi: 10.1016/j.mib.2021.11.005. Epub 2021 Nov 29. Curr Opin Microbiol. 2022. PMID: 34856509 Review.
-
Identification of novel, cryptic Clostridioides species isolates from environmental samples collected from diverse geographical locations.Microb Genom. 2022 Feb;8(2):000742. doi: 10.1099/mgen.0.000742. Microb Genom. 2022. PMID: 35166655 Free PMC article.
-
The analysis of para-cresol production and tolerance in Clostridium difficile 027 and 012 strains.BMC Microbiol. 2011 Apr 28;11:86. doi: 10.1186/1471-2180-11-86. BMC Microbiol. 2011. PMID: 21527013 Free PMC article.
-
Pathogenicity and virulence of Clostridioides difficile.Virulence. 2023 Dec;14(1):2150452. doi: 10.1080/21505594.2022.2150452. Virulence. 2023. PMID: 36419222 Free PMC article. Review.
Cited by
-
Comparative genome analyses of clinical and non-clinical Clostridioides difficile strains.Front Microbiol. 2024 Jun 27;15:1404491. doi: 10.3389/fmicb.2024.1404491. eCollection 2024. Front Microbiol. 2024. PMID: 38993487 Free PMC article.
References
-
- Karaoui WR, Rustam LBO, Bou Daher H, Rimmani HH, Rasheed SS, Matar GM, Mahfouz R, Araj GF, Zahreddine N, Kanj SS, Berger FK, Gärtner B, El Sabbagh R, Sharara AI. 2020. Incidence, outcome, and risk factors for recurrence of nosocomial Clostridioides difficile infection in adults: a prospective cohort study. J Infect Public Health 13:485–490. doi: 10.1016/j.jiph.2019.11.005 - DOI - PubMed
-
- Du T, Choi KB, Silva A, Golding GR, Pelude L, Hizon R, Al-Rawahi GN, Brooks J, Chow B, Collet JC, et al. 2022. Characterization of healthcare-associated and community-associated Clostridioides difficile infections among adults, Canada, 2015–2019. Emerg Infect Dis 28:1128–1136. doi: 10.3201/eid2806.212262 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

