PIEZO1 is essential for the survival and proliferation of acute myeloid leukemia cells
- PMID: 38334477
- PMCID: PMC10854442
- DOI: 10.1002/cam4.6984
PIEZO1 is essential for the survival and proliferation of acute myeloid leukemia cells
Abstract
Introduction: Leukemogenesis is a complex process that interconnects tumoral cells with their microenvironment, but the effect of mechanosensing in acute myeloid leukemia (AML) blasts is poorly known. PIEZO1 perceives and transmits the constraints of the environment to human cells by acting as a non-selective calcium channel, but very little is known about its role in leukemogenesis.
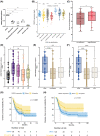
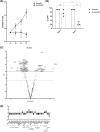
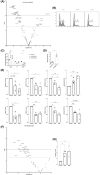
Results: For the first time, we show that PIEZO1 is preferentially expressed in healthy hematopoietic stem and progenitor cells in human hematopoiesis, and globally overexpressed in AML cells. In AML subtypes, PIEZO1 expression associates with favorable outcomes as better overall (OS) and disease-free survival (DFS). If PIEZO1 is expressed and functional in THP1 leukemic myeloid cell line, its chemical activation doesn't impact the proliferation, differentiation, nor survival of cells. However, the downregulation of PIEZO1 expression dramatically reduces the proliferation and the survival of THP1 cells. We show that PIEZO1 knock-down blocks the cell cycle in G0/G1 phases of AML cells, impairs the DNA damage response pathways, and critically increases cell death by triggering extrinsic apoptosis pathways.
Conclusions: Altogether, our results reveal a new role for PIEZO1 mechanosensing in the survival and proliferation of leukemic blasts, which could pave the way for new therapeutic strategies to target AML cells.
Keywords: PIEZO1; acute myeloid leukemia; apoptosis; cell cycle; proliferation; survival.
© 2024 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

