Postnatal Dynamic Ciliary ARL13B and ADCY3 Localization in the Mouse Brain
- PMID: 38334651
- PMCID: PMC10854790
- DOI: 10.3390/cells13030259
Postnatal Dynamic Ciliary ARL13B and ADCY3 Localization in the Mouse Brain
Abstract
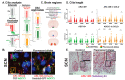
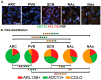
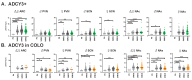
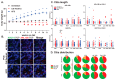
Primary cilia are hair-like structures found on nearly all mammalian cell types, including cells in the developing and adult brain. A diverse set of receptors and signaling proteins localize within cilia to regulate many physiological and developmental pathways, including the Hedgehog (Hh) pathway. Defects in cilia structure, protein localization, and function lead to genetic disorders called ciliopathies, which present with various clinical features that include several neurodevelopmental phenotypes and hyperphagia-associated obesity. Despite their dysfunction being implicated in several disease states, understanding their roles in central nervous system (CNS) development and signaling has proven challenging. We hypothesize that dynamic changes to ciliary protein composition contribute to this challenge and may reflect unrecognized diversity of CNS cilia. The proteins ARL13B and ADCY3 are established markers of cilia in the brain. ARL13B is a regulatory GTPase important for regulating cilia structure, protein trafficking, and Hh signaling, and ADCY3 is a ciliary adenylyl cyclase. Here, we examine the ciliary localization of ARL13B and ADCY3 in the perinatal and adult mouse brain. We define changes in the proportion of cilia enriched for ARL13B and ADCY3 depending on brain region and age. Furthermore, we identify distinct lengths of cilia within specific brain regions of male and female mice. ARL13B+ cilia become relatively rare with age in many brain regions, including the hypothalamic feeding centers, while ADCY3 becomes a prominent cilia marker in the mature adult brain. It is important to understand the endogenous localization patterns of these proteins throughout development and under different physiological conditions as these common cilia markers may be more dynamic than initially expected. Understanding regional- and developmental-associated cilia protein composition signatures and physiological condition cilia dynamic changes in the CNS may reveal the molecular mechanisms associated with the features commonly observed in ciliopathy models and ciliopathies, like obesity and diabetes.
Keywords: ADCY3; ARL13B; brain; cilia; energy homeostasis; hypothalamus; nucleus accumbens.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
- R35GM148416/GM/NIGMS NIH HHS/United States
- R35GM122549/GM/NIGMS NIH HHS/United States
- R01 DK124769/DK/NIDDK NIH HHS/United States
- R01DK060540/DK/NIDDK NIH HHS/United States
- R35 GM122549/GM/NIGMS NIH HHS/United States
- R01DK106404/DK/NIDDK NIH HHS/United States
- P30 DK098722/DK/NIDDK NIH HHS/United States
- R01 DK060540/DK/NIDDK NIH HHS/United States
- R01 DK106404/DK/NIDDK NIH HHS/United States
- R01 NS090029/NS/NINDS NIH HHS/United States
- R35 GM148416/GM/NIGMS NIH HHS/United States
- R01DK114008/DK/NIDDK NIH HHS/United States
- R01NS090029/NH/NIH HHS/United States
- R01DK124769/DK/NIDDK NIH HHS/United States
- R01 DK114008/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

