BubR1 controls starvation-induced lipolysis via IMD signaling pathway in Drosophila
- PMID: 38334966
- PMCID: PMC10929803
- DOI: 10.18632/aging.205533
BubR1 controls starvation-induced lipolysis via IMD signaling pathway in Drosophila
Abstract
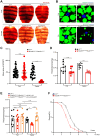
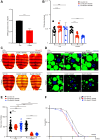
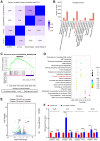
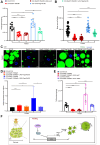
Lipolysis, the key process releasing fat acids to generate energy in adipose tissues, correlates with starvation resistance. Nevertheless, its detail mechanisms remain elusive. BubR1, an essential mitotic regulator, ensures proper chromosome alignment and segregation during mitosis, but its physiological functions are largely unknown. Here, we use Drosophila adult fat body, the major lipid storage organ, to study the functions of BubR1 in lipolysis. We show that both whole body- and fat body-specific BubR1 depletions increase lipid degradation and shorten the lifespan under fasting but not feeding. Relish, the conserved regulator of IMD signaling pathway, acts as the downstream target of BubR1 to control the expression level of Bmm and modulate the lipolysis upon fasting. Thus, our study reveals new functions of BubR1 in starvation-induced lipolysis and provides new insights into the molecular mechanisms of lipolysis mediated by IMD signaling pathway.
Keywords: Bmm; BubR1; Relish; immunity and metabolism; lipolysis; metabolic adaptation.
Conflict of interest statement
Figures
Similar articles
-
NF-κB Shapes Metabolic Adaptation by Attenuating Foxo-Mediated Lipolysis in Drosophila.Dev Cell. 2019 Jun 3;49(5):802-810.e6. doi: 10.1016/j.devcel.2019.04.009. Epub 2019 May 9. Dev Cell. 2019. PMID: 31080057 Free PMC article.
-
Hedgehog signaling promotes lipolysis in adipose tissue through directly regulating Bmm/ATGL lipase.Dev Biol. 2020 Jan 1;457(1):128-139. doi: 10.1016/j.ydbio.2019.09.009. Epub 2019 Sep 21. Dev Biol. 2020. PMID: 31550483 Free PMC article.
-
Bub1 and Bub3 regulate metabolic adaptation via macrolipophagy in Drosophila.Cell Rep. 2023 Apr 25;42(4):112343. doi: 10.1016/j.celrep.2023.112343. Epub 2023 Apr 5. Cell Rep. 2023. PMID: 37027296
-
The spindle checkpoint proteins BUB1 and BUBR1: (SLiM)ming down to the basics.Trends Biochem Sci. 2022 Apr;47(4):352-366. doi: 10.1016/j.tibs.2022.01.004. Epub 2022 Feb 17. Trends Biochem Sci. 2022. PMID: 35184951 Review.
-
Beyond immunity: The Imd pathway as a coordinator of host defense, organismal physiology and behavior.Dev Comp Immunol. 2018 Jun;83:51-59. doi: 10.1016/j.dci.2017.11.008. Epub 2017 Nov 14. Dev Comp Immunol. 2018. PMID: 29146454 Review.
Cited by
-
Novel insights into the circadian modulation of lipid metabolism in chicken livers revealed by RNA sequencing and weighted gene co-expression network analysis.Poult Sci. 2024 Dec;103(12):104321. doi: 10.1016/j.psj.2024.104321. Epub 2024 Sep 16. Poult Sci. 2024. PMID: 39361997 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

