Functional EPAS1/ HIF2A missense variant is associated with hematocrit in Andean highlanders
- PMID: 38335297
- PMCID: PMC10857371
- DOI: 10.1126/sciadv.adj5661
Functional EPAS1/ HIF2A missense variant is associated with hematocrit in Andean highlanders
Abstract
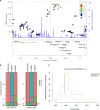
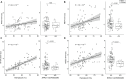
Hypoxia-inducible factor pathway genes are linked to adaptation in both human and nonhuman highland species. EPAS1, a notable target of hypoxia adaptation, is associated with relatively lower hemoglobin concentration in Tibetans. We provide evidence for an association between an adaptive EPAS1 variant (rs570553380) and the same phenotype of relatively low hematocrit in Andean highlanders. This Andean-specific missense variant is present at a modest frequency in Andeans and absent in other human populations and vertebrate species except the coelacanth. CRISPR-base-edited human cells with this variant exhibit shifts in hypoxia-regulated gene expression, while metabolomic analyses reveal both genotype and phenotype associations and validation in a lowland population. Although this genocopy of relatively lower hematocrit in Andean highlanders parallels well-replicated findings in Tibetans, it likely involves distinct pathway responses based on a protein-coding versus noncoding variants, respectively. These findings illuminate how unique variants at EPAS1 contribute to the same phenotype in Tibetans and a subset of Andean highlanders despite distinct evolutionary trajectories.
Figures


References
-
- Beall C. M., Cavalleri G. L., Deng L., Elston R. C., Gao Y., Knight J., Li C., Li J. C., Liang Y., McCormack M., Montgomery H. E., Pan H., Robbins P. A., Shianna K. V., Tam S. C., Tsering N., Veeramah K. R., Wang W., Wangdui P., Weale M. E., Xu Y., Xu Z., Yang L., Zaman M. J., Zeng C., Zhang L., Zhang X., Zhaxi P., Zheng Y. T., Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107, 11459–11464 (2010). - PMC - PubMed
-
- Simonson T. S., Yang Y., Huff C. D., Yun H., Qin G., Witherspoon D. J., Bai Z., Lorenzo F. R., Xing J., Jorde L. B., Prchal J. T., Ge R., Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 (2010). - PubMed
-
- Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z. X. P., Pool J. E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T. S., Zheng H., Liu T., He W., Li K., Luo R., Nie X., Wu H., Zhao M., Cao H., Zou J., Shan Y., Li S., Yang Q., Asan, Ni P., Tian G., Xu J., Liu X., Jiang T., Wu R., Zhou G., Tang M., Qin J., Wang T., Feng S., Li G., Huasang, Luosang J., Wang W., Chen F., Wang Y., Zheng X., Li Z., Bianba Z., Yang G., Wang X., Tang S., Gao G., Chen Y., Luo Z., Gusang L., Cao Z., Zhang Q., Ouyang W., Ren X., Liang H., Zheng H., Huang Y., Li J., Bolund L., Kristiansen K., Li Y., Zhang Y., Zhang X., Li R., Li S., Yang H., Nielsen R., Wang J., Wang J., Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). - PMC - PubMed

