Single-Cell Analysis Identifies NOTCH3-Mediated Interactions between Stromal Cells That Promote Microenvironment Remodeling and Invasion in Lung Adenocarcinoma
- PMID: 38335304
- PMCID: PMC11063690
- DOI: 10.1158/0008-5472.CAN-23-1183
Single-Cell Analysis Identifies NOTCH3-Mediated Interactions between Stromal Cells That Promote Microenvironment Remodeling and Invasion in Lung Adenocarcinoma
Abstract
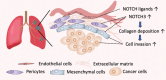
Cancer immunotherapy has revolutionized the treatment of lung adenocarcinoma (LUAD); however, a significant proportion of patients do not respond. Recent transcriptomic studies to understand determinants of immunotherapy response have pinpointed stromal-mediated resistance mechanisms. To gain a better understanding of stromal biology at the cellular and molecular level in LUAD, we performed single-cell RNA sequencing of 256,379 cells, including 13,857 mesenchymal cells, from 9 treatment-naïve patients. Among the mesenchymal cell subsets, FAP+PDPN+ cancer-associated fibroblasts (CAF) and ACTA2+MCAM+ pericytes were enriched in tumors and differentiated from lung-resident fibroblasts. Imaging mass cytometry revealed that both subsets were topographically adjacent to the perivascular niche and had close spatial interactions with endothelial cells (EC). Modeling of ligand and receptor interactomes between mesenchymal and ECs identified that NOTCH signaling drives these cell-to-cell interactions in tumors, with pericytes and CAFs as the signal receivers and arterial and PLVAPhigh immature neovascular ECs as the signal senders. Either pharmacologically blocking NOTCH signaling or genetically depleting NOTCH3 levels in mesenchymal cells significantly reduced collagen production and suppressed cell invasion. Bulk RNA sequencing data demonstrated that NOTCH3 expression correlated with poor survival in stroma-rich patients and that a T cell-inflamed gene signature only predicted survival in patients with low NOTCH3. Collectively, this study provides valuable insights into the role of NOTCH3 in regulating tumor stroma biology, warranting further studies to elucidate the clinical implications of targeting NOTCH3 signaling.
Significance: NOTCH3 signaling activates tumor-associated mesenchymal cells, increases collagen production, and augments cell invasion in lung adenocarcinoma, suggesting its critical role in remodeling tumor stroma.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Myers DJ, Wallen JM. Lung adenocarcinoma. StatPearls. Treasure Island, FL;2023.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

