Hypoxia causes trophoblast cell ferroptosis to induce miscarriage through lnc-HZ06/HIF1α-SUMO/NCOA4 axis
- PMID: 38335622
- PMCID: PMC10869313
- DOI: 10.1016/j.redox.2024.103073
Hypoxia causes trophoblast cell ferroptosis to induce miscarriage through lnc-HZ06/HIF1α-SUMO/NCOA4 axis
Abstract
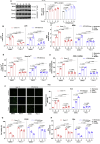
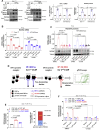
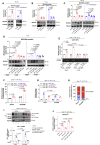
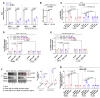
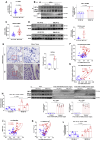
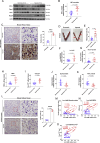
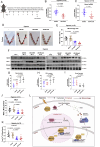
Defects of human trophoblast cells may induce miscarriage (abnormal early embryo loss), which is generally regulated by lncRNAs. Ferroptosis is a newly identified iron-dependent programmed cell death. Hypoxia is an important and unavoidable feature in mammalian cells. However, whether hypoxia might induce trophoblast cell ferroptosis and then induce miscarriage, as well as regulated by a lncRNA, was completely unknown. In this work, we discovered at the first time that hypoxia could result in ferroptosis of human trophoblast cells and then induce miscarriage. We also identified a novel lncRNA (lnc-HZ06) that simultaneously regulated hypoxia (indicated by HIF1α protein), ferroptosis, and miscarriage. In mechanism, HIF1α-SUMO, instead of HIF1α itself, primarily acted as a transcription factor to promote the transcription of NCOA4 (ferroptosis indicator) in hypoxic trophoblast cells. Lnc-HZ06 promoted the SUMOylation of HIF1α by suppressing SENP1-mediated deSUMOylation. HIF1α-SUMO also acted as a transcription factor to promote lnc-HZ06 transcription. Thus, both lnc-HZ06 and HIF1α-SUMO formed a positive auto-regulatory feedback loop. This loop was up-regulated in hypoxic trophoblast cells, in RM villous tissues, and in placental tissues of hypoxia-treated mice, which further induced ferroptosis and miscarriage by up-regulating HIF1α-SUMO-mediated NCOA4 transcription. Furthermore, knockdown of either murine lnc-hz06 or Ncoa4 could efficiently suppress ferroptosis and alleviate miscarriage in hypoxic mouse model. Taken together, this study provided new insights in understanding the regulatory roles of lnc-HZ06/HIF1α-SUMO/NCOA4 axis among hypoxia, ferroptosis, and miscarriage, and also offered an effective approach for treatment against miscarriage.
Keywords: Hypoxia; LncRNA or lnc-HZ06; Recurrent miscarriage; SUMOylated HIF1α and NCOA4; Trophoblast cell ferroptosis.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest None.
Figures







References
-
- Rai R., Regan L. Recurrent miscarriage. Lancet. 2006;368(9535):601–611. - PubMed
-
- Green D.M., O'Donoghue K. A review of reproductive outcomes of women with two consecutive miscarriages and no living child. J. Obstet. Gynaecol. 2019;39(6):816–821. - PubMed
-
- Basile N., Elkhatib I., Meseguer M. A strength, weaknesses, opportunities and threats analysis on time lapse. Curr. Opin. Obstet. Gynecol. 2019;31(3):148–155. - PubMed
Further reading
-
- Mi C., et al. Lnc-HZ05 regulates BPDE-inhibited human trophoblast cell proliferation and affects the occurrence of miscarriage by directly binding with miR-hz05. Cell Biol. Toxicol. 2022;38(6):1137–1157. - PubMed
-
- Xu Z., et al. Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci. Total Environ. 2021;776 - PubMed
-
- Liang T., et al. Novel lnc-HZ03 and miR-hz03 promote BPDE-induced human trophoblastic cell apoptosis and induce miscarriage by upregulating p53/SAT1 pathway. Cell Biol. Toxicol. 2021;37(6):951–970. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

