Integrative high-throughput enhancer surveying and functional verification divulges a YY2-condensed regulatory axis conferring risk for osteoporosis
- PMID: 38335956
- PMCID: PMC10943593
- DOI: 10.1016/j.xgen.2024.100501
Integrative high-throughput enhancer surveying and functional verification divulges a YY2-condensed regulatory axis conferring risk for osteoporosis
Abstract
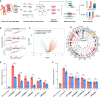
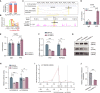
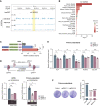
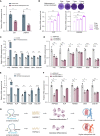
The precise roles of chromatin organization at osteoporosis risk loci remain largely elusive. Here, we combined chromatin interaction conformation (Hi-C) profiling and self-transcribing active regulatory region sequencing (STARR-seq) to qualify enhancer activities of prioritized osteoporosis-associated single-nucleotide polymorphisms (SNPs). We identified 319 SNPs with biased allelic enhancer activity effect (baaSNPs) that linked to hundreds of candidate target genes through chromatin interactions across 146 loci. Functional characterizations revealed active epigenetic enrichment for baaSNPs and prevailing osteoporosis-relevant regulatory roles for their chromatin interaction genes. Further motif enrichment and network mapping prioritized several putative, key transcription factors (TFs) controlling osteoporosis binding to baaSNPs. Specifically, we selected one top-ranked TF and deciphered that an intronic baaSNP (rs11202530) could allele-preferentially bind to YY2 to augment PAPSS2 expression through chromatin interactions and promote osteoblast differentiation. Our results underline the roles of TF-mediated enhancer-promoter contacts for osteoporosis, which may help to better understand the intricate molecular regulatory mechanisms underlying osteoporosis risk loci.
Keywords: Hi-C; PAPSS2; STARR-seq; YY2; chromatin interaction; enhancer; genetic regulatory network; osteoblast differentiation; osteoporosis; transcription factor.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
High-throughput functional dissection of noncoding SNPs with biased allelic enhancer activity for insulin resistance-relevant phenotypes.Am J Hum Genet. 2023 Aug 3;110(8):1266-1288. doi: 10.1016/j.ajhg.2023.07.002. Epub 2023 Jul 27. Am J Hum Genet. 2023. PMID: 37506691 Free PMC article.
-
Multiple Functional Variants at 13q14 Risk Locus for Osteoporosis Regulate RANKL Expression Through Long-Range Super-Enhancer.J Bone Miner Res. 2018 Jul;33(7):1335-1346. doi: 10.1002/jbmr.3419. Epub 2018 May 17. J Bone Miner Res. 2018. PMID: 29528523
-
An Osteoporosis Risk SNP at 1p36.12 Acts as an Allele-Specific Enhancer to Modulate LINC00339 Expression via Long-Range Loop Formation.Am J Hum Genet. 2018 May 3;102(5):776-793. doi: 10.1016/j.ajhg.2018.03.001. Epub 2018 Apr 26. Am J Hum Genet. 2018. PMID: 29706346 Free PMC article.
-
Functional genomic assays to annotate enhancer-promoter interactions genome wide.Hum Mol Genet. 2022 Oct 20;31(R1):R97-R104. doi: 10.1093/hmg/ddac204. Hum Mol Genet. 2022. PMID: 36018818 Free PMC article. Review.
-
Principle and application of self-transcribing active regulatory region sequencing in enhancer discovery research.Yi Chuan. 2024 Aug;46(8):589-602. doi: 10.16288/j.yczz.24-149. Yi Chuan. 2024. PMID: 39140141 Review.
Cited by
-
RUNX2 Phase Separation Mediates Long-Range Regulation Between Osteoporosis-Susceptibility Variant and XCR1 to Promote Osteoblast Differentiation.Adv Sci (Weinh). 2025 Feb;12(6):e2413561. doi: 10.1002/advs.202413561. Epub 2024 Dec 20. Adv Sci (Weinh). 2025. PMID: 39704037 Free PMC article.
-
Comprehensive Analysis of the Role of Metabolic Features in Osteoporosis: A Multi-Omics Analysis.Int J Gen Med. 2025 May 26;18:2727-2739. doi: 10.2147/IJGM.S515717. eCollection 2025. Int J Gen Med. 2025. PMID: 40453197 Free PMC article.
-
Genetic transcriptional regulation profiling of cartilage reveals pathogenesis of osteoarthritis.EBioMedicine. 2025 Jul;117:105821. doi: 10.1016/j.ebiom.2025.105821. Epub 2025 Jun 26. EBioMedicine. 2025. PMID: 40577937 Free PMC article.
-
Analysis of biased allelic enhancer activity of schizophrenia-linked common variants.Commun Biol. 2025 Jul 10;8(1):1034. doi: 10.1038/s42003-025-08456-3. Commun Biol. 2025. PMID: 40640362 Free PMC article.
References
-
- Trajanoska K., Morris J.A., Oei L., Zheng H.F., Evans D.M., Kiel D.P., Ohlsson C., Richards J.B., Rivadeneira F., GEFOS/GENOMOS consortium and the 23andMe research team Assessment of the genetic and clinical determinants of fracture risk: genome wide association and mendelian randomisation study. BMJ. 2018;362:k3225. - PMC - PubMed
-
- Medina-Gomez C., Kemp J.P., Dimou N.L., Kreiner E., Chesi A., Zemel B.S., Bønnelykke K., Boer C.G., Ahluwalia T.S., Bisgaard H., et al. Bivariate genome-wide association meta-analysis of pediatric musculoskeletal traits reveals pleiotropic effects at the SREBF1/TOM1L2 locus. Nat. Commun. 2017;8:121. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

