Interorgan rhythmicity as a feature of healthful metabolism
- PMID: 38335957
- PMCID: PMC10990795
- DOI: 10.1016/j.cmet.2024.01.009
Interorgan rhythmicity as a feature of healthful metabolism
Abstract
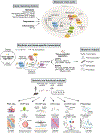
The finding that animals with circadian gene mutations exhibit diet-induced obesity and metabolic syndrome with hypoinsulinemia revealed a distinct role for the clock in the brain and peripheral tissues. Obesogenic diets disrupt rhythmic sleep/wake patterns, feeding behavior, and transcriptional networks, showing that metabolic signals reciprocally control the clock. Providing access to high-fat diet only during the sleep phase (light period) in mice accelerates weight gain, whereas isocaloric time-restricted feeding during the active period enhances energy expenditure due to circadian induction of adipose thermogenesis. This perspective focuses on advances and unanswered questions in understanding the interorgan circadian control of healthful metabolism.
Keywords: circadian; diabetes; epigenetics; insulin; metabolism; molecular clock; obesity; sleep; thermogenesis; transcription.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The author declares no competing interests.
Figures


Similar articles
-
Short-term feeding at the wrong time is sufficient to desynchronize peripheral clocks and induce obesity with hyperphagia, physical inactivity and metabolic disorders in mice.Metabolism. 2016 May;65(5):714-727. doi: 10.1016/j.metabol.2016.02.003. Epub 2016 Feb 9. Metabolism. 2016. PMID: 27085778
-
Timing of food intake is more potent than habitual voluntary exercise to prevent diet-induced obesity in mice.Chronobiol Int. 2019 Jan;36(1):57-74. doi: 10.1080/07420528.2018.1516672. Epub 2018 Sep 13. Chronobiol Int. 2019. PMID: 30212233
-
Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock.Cell Metab. 2019 Feb 5;29(2):303-319.e4. doi: 10.1016/j.cmet.2018.08.004. Epub 2018 Aug 30. Cell Metab. 2019. PMID: 30174302 Free PMC article.
-
Circadian mechanisms in adipose tissue bioenergetics and plasticity.Genes Dev. 2023 Jun 1;37(11-12):454-473. doi: 10.1101/gad.350759.123. Epub 2023 Jun 26. Genes Dev. 2023. PMID: 37364987 Free PMC article. Review.
-
Interactions between metabolism and circadian clocks: reciprocal disturbances.Ann N Y Acad Sci. 2011 Dec;1243:30-46. doi: 10.1111/j.1749-6632.2011.06246.x. Ann N Y Acad Sci. 2011. PMID: 22211891 Review.
Cited by
-
Chronic mistimed feeding results in renal fibrosis and disrupted circadian blood pressure rhythms.Am J Physiol Renal Physiol. 2024 Nov 1;327(5):F683-F696. doi: 10.1152/ajprenal.00047.2024. Epub 2024 Aug 29. Am J Physiol Renal Physiol. 2024. PMID: 39205662
-
Feeding disruptions lead to a significant increase in disease modules in adult mice.Heliyon. 2025 Jan 7;11(2):e41774. doi: 10.1016/j.heliyon.2025.e41774. eCollection 2025 Jan 30. Heliyon. 2025. PMID: 39882459 Free PMC article.
-
Circadian rhythm disruption: a potential trigger in Parkinson's disease pathogenesis.Front Cell Neurosci. 2024 Oct 30;18:1464595. doi: 10.3389/fncel.2024.1464595. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39539340 Free PMC article. Review.
-
Scheduled feeding improves sleep in a mouse model of Huntington's disease.Front Neurosci. 2024 Aug 5;18:1427125. doi: 10.3389/fnins.2024.1427125. eCollection 2024. Front Neurosci. 2024. PMID: 39161652 Free PMC article.
-
Scheduled feeding improves sleep in a mouse model of Huntington's disease.bioRxiv [Preprint]. 2024 May 7:2024.05.04.592428. doi: 10.1101/2024.05.04.592428. bioRxiv. 2024. Update in: Front Neurosci. 2024 Aug 05;18:1427125. doi: 10.3389/fnins.2024.1427125. PMID: 38766112 Free PMC article. Updated. Preprint.
References
-
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, and Schibler U (2002). The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. S0092867402008255 [pii]. - PubMed
-
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, and Hogenesch JB (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320. S0092867402007225 [pii]. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

