NY-ESO-1-specific T cell receptor-engineered T cells and Tranilast, a TRPV2 antagonist bivalent treatment enhances the killing of esophageal cancer: a dual-targeted cancer therapeutic route
- PMID: 38336680
- PMCID: PMC10858587
- DOI: 10.1186/s12935-024-03249-w
NY-ESO-1-specific T cell receptor-engineered T cells and Tranilast, a TRPV2 antagonist bivalent treatment enhances the killing of esophageal cancer: a dual-targeted cancer therapeutic route
Abstract
Background: Esophageal cancer (EC) is a global canker notorious for causing high mortality due to its relentless incidence rate, convoluted with unyielding recurrence and metastasis. However, these intricacies of EC are associated with an immoderate expression of NY-ESO-1 antigen, presenting a lifeline for adoptive T cell therapy. We hypothesized that naturally isolated higher-affinity T cell receptors (TCRs) that bind to NY-ESO-1 would allow T lymphocytes to target EC with a pronounced antitumor response efficacy. Also, targeting TRPV2, which is associated with tumorigenesis in EC, creates an avenue for dual-targeted therapy. We exploited the dual-targeting antitumor efficacy against EC.
Methods: We isolated antigen-specific TCRs (asTCRs) from a naive library constructed with TCRs obtained from enriched cytotoxic T lymphocytes. The robustness of our asTCRs and their TCR-T cell derivatives, Tranilast (TRPV2 inhibitor), and their bivalent treatment were evaluated with prospective cross-reactive human-peptide variants and tumor cells.
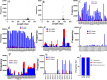
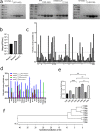
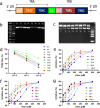
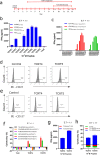
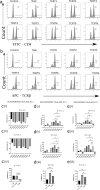
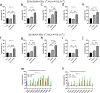
Results: Our study demonstrated that our naive unenhanced asTCRs and their TCR-Ts perpetuated their cognate HLA-A*02:01/NY-ESO-1(157-165) specificity, killing varying EC cells with higher cytotoxicity compared to the known affinity-enhanced TCR (TCRe) and its wild-type (TCR0) which targets the same NY-ESO-1 antigen. Furthermore, the TCR-Ts and Tranilast bivalent treatment showed superior EC killing compared to any of their monovalent treatments of either TCR-T or Tranilast.
Conclusion: Our findings suggest that dual-targeted immunotherapy may have a superior antitumor effect. Our study presents a technique to evolve novel, robust, timely therapeutic strategies and interventions for EC and other malignancies.
Keywords: Adoptive therapy; Cytotoxic T lymphocytes; Esophageal cancer (EC); NY-ESO-1 antigen; T cell receptor-engineered T cell (TCR-T); Tranilast.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

