Androgen deprivation induces double-null prostate cancer via aberrant nuclear export and ribosomal biogenesis through HGF and Wnt activation
- PMID: 38336745
- PMCID: PMC10858246
- DOI: 10.1038/s41467-024-45489-4
Androgen deprivation induces double-null prostate cancer via aberrant nuclear export and ribosomal biogenesis through HGF and Wnt activation
Abstract
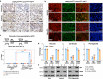
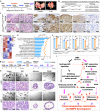
Androgen deprivation therapy (ADT) targeting androgen/androgen receptor (AR)- signaling pathways is the main therapy for advanced prostate cancer (PCa). However, ADT eventually fails in most patients who consequently develop castration-resistant prostate cancer (CRPC). While more potent AR antagonists and blockers for androgen synthesis were developed to improve clinical outcomes, they also show to induce more diverse CRPC phenotypes. Specifically, the AR- and neuroendocrine-null PCa, DNPC, occurs in abiraterone and enzalutamide-treated patients. Here, we uncover that current ADT induces aberrant HGF/MET signaling activation that further elevates Wnt/β-catenin signaling in human DNPC samples. Co-activation of HGF/MET and Wnt/β-catenin axes in mouse prostates induces DNPC-like lesions. Single-cell RNA sequencing analyses identify increased expression and activity of XPO1 and ribosomal proteins in mouse DNPC-like cells. Elevated expression of XPO1 and ribosomal proteins is also identified in clinical DNPC specimens. Inhibition of XPO1 and ribosomal pathways represses DNPC growth in both in vivo and ex vivo conditions, evidencing future therapeutic targets.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Jenster G. The role of the androgen receptor in the development and progression of prostate cancer. Semin. Oncol. 1999;26:407–421. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

