Large-scale integrative analysis of juvenile idiopathic arthritis for new insight into its pathogenesis
- PMID: 38336809
- PMCID: PMC10858498
- DOI: 10.1186/s13075-024-03280-2
Large-scale integrative analysis of juvenile idiopathic arthritis for new insight into its pathogenesis
Abstract
Background: Juvenile idiopathic arthritis (JIA) is one of the most prevalent rheumatic disorders in children and is classified as an autoimmune disease (AID). While a robust genetic contribution to JIA etiology has been established, the exact pathogenesis remains unclear.
Methods: To prioritize biologically interpretable susceptibility genes and proteins for JIA, we conducted transcriptome-wide and proteome-wide association studies (TWAS/PWAS). Then, to understand the genetic architecture of JIA, we systematically analyzed single-nucleotide polymorphism (SNP)-based heritability, a signature of natural selection, and polygenicity. Next, we conducted HLA typing using multi-ethnicity RNA sequencing data. Additionally, we examined the T cell receptor (TCR) repertoire at a single-cell level to explore the potential links between immunity and JIA risk.
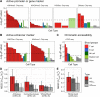
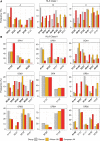
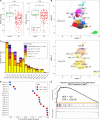
Results: We have identified 19 TWAS genes and two PWAS proteins associated with JIA risks. Furthermore, we observe that the heritability and cell type enrichment analysis of JIA are enriched in T lymphocytes and HLA regions and that JIA shows higher polygenicity compared to other AIDs. In multi-ancestry HLA typing, B*45:01 is more prevalent in African JIA patients than in European JIA patients, whereas DQA1*01:01, DQA1*03:01, and DRB1*04:01 exhibit a higher frequency in European JIA patients. Using single-cell immune repertoire analysis, we identify clonally expanded T cell subpopulations in JIA patients, including CXCL13+BHLHE40+ TH cells which are significantly associated with JIA risks.
Conclusion: Our findings shed new light on the pathogenesis of JIA and provide a strong foundation for future mechanistic studies aimed at uncovering the molecular drivers of JIA.
Keywords: Juvenile idiopathic arthritis; Multi-ethnicity RNA typing; T cell receptor (TCR) repertoire; Transcriptome-wide and proteome-wide association studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

