Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa
- PMID: 38336881
- PMCID: PMC10914600
- DOI: 10.1038/s41564-024-01603-2
Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa
Abstract
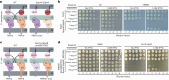
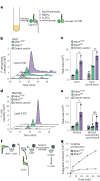
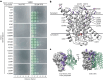
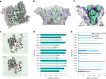
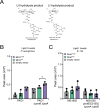
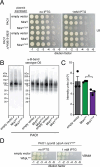
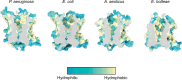
Many bacterial surface glycans such as the peptidoglycan (PG) cell wall are built from monomeric units linked to a polyprenyl lipid carrier. How this limiting carrier is distributed among competing pathways has remained unclear. Here we describe the isolation of hyperactive variants of Pseudomonas aeruginosa MraY, the enzyme that forms the first lipid-linked PG precursor. These variants result in the elevated production of the final PG precursor lipid II in cells and are hyperactive in vitro. The activated MraY variants have substitutions that map to a cavity on the extracellular side of the dimer interface, far from the active site. Our structural and molecular dynamics results suggest that this cavity is a binding site for externalized lipid II. Overall, our results support a model in which excess externalized lipid II allosterically inhibits MraY, providing a feedback mechanism that prevents the sequestration of lipid carrier in the PG biogenesis pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
A feedback control mechanism governs the synthesis of lipid-linked precursors of the bacterial cell wall.bioRxiv [Preprint]. 2023 Aug 1:2023.08.01.551478. doi: 10.1101/2023.08.01.551478. bioRxiv. 2023. Update in: Nat Microbiol. 2024 Mar;9(3):763-775. doi: 10.1038/s41564-024-01603-2. PMID: 37577621 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

