Short tandem repeat mutations regulate gene expression in colorectal cancer
- PMID: 38336885
- PMCID: PMC10858039
- DOI: 10.1038/s41598-024-53739-0
Short tandem repeat mutations regulate gene expression in colorectal cancer
Abstract
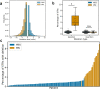
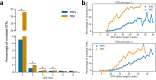
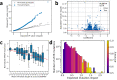
Short tandem repeat (STR) mutations are prevalent in colorectal cancer (CRC), especially in tumours with the microsatellite instability (MSI) phenotype. While STR length variations are known to regulate gene expression under physiological conditions, the functional impact of STR mutations in CRC remains unclear. Here, we integrate STR mutation data with clinical information and gene expression data to study the gene regulatory effects of STR mutations in CRC. We confirm that STR mutability in CRC highly depends on the MSI status, repeat unit size, and repeat length. Furthermore, we present a set of 1244 putative expression STRs (eSTRs) for which the STR length is associated with gene expression levels in CRC tumours. The length of 73 eSTRs is associated with expression levels of cancer-related genes, nine of which are CRC-specific genes. We show that linear models describing eSTR-gene expression relationships allow for predictions of gene expression changes in response to eSTR mutations. Moreover, we found an increased mutability of eSTRs in MSI tumours. Our evidence of gene regulatory roles for eSTRs in CRC highlights a mostly overlooked way through which tumours may modulate their phenotypes. Future extensions of these findings could uncover new STR-based targets in the treatment of cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

